bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1849_orf2
Length=136
Score E
Sequences producing significant alignments: (Bits) Value
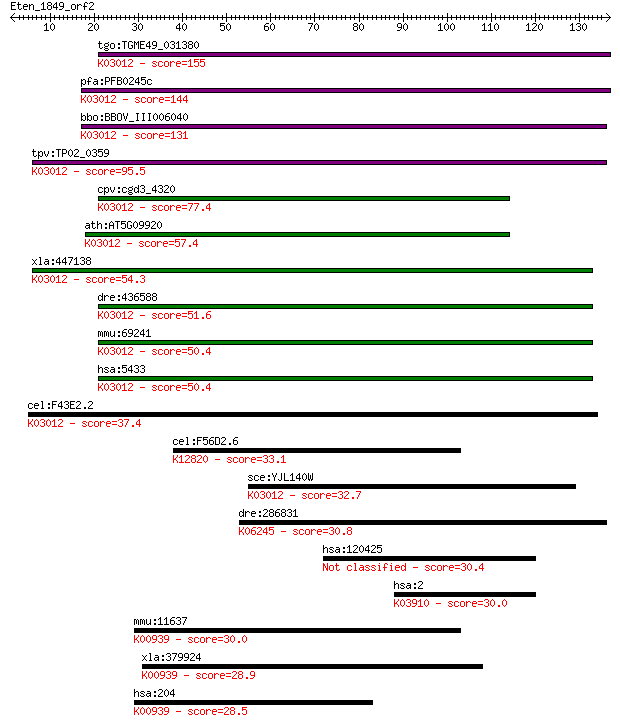
tgo:TGME49_031380 DNA-directed RNA polymerase II, putative ; K... 155 3e-38
pfa:PFB0245c DNA-directed RNA polymerase II 16 kDa subunit, pu... 144 1e-34
bbo:BBOV_III006040 17.m07533; RNA polymerase Rpb4 family prote... 131 7e-31
tpv:TP02_0359 DNA-directed RNA polymerase II subunit Rpb4; K03... 95.5 4e-20
cpv:cgd3_4320 hypothetical protein ; K03012 DNA-directed RNA p... 77.4 1e-14
ath:AT5G09920 NRPB4; NRPB4; DNA-directed RNA polymerase; K0301... 57.4 1e-08
xla:447138 polr2d, MGC85403; polymerase (RNA) II (DNA directed... 54.3 1e-07
dre:436588 polr2d, wu:fa95b11, zgc:110435; polymerase (RNA) II... 51.6 8e-07
mmu:69241 Polr2d, 2310002G05Rik, 2610028L19Rik, HSRBP4, RBP4; ... 50.4 2e-06
hsa:5433 POLR2D, HSRBP4, HSRPB4, RBP4, RPB16; polymerase (RNA)... 50.4 2e-06
cel:F43E2.2 rpb-4; RNA Polymerase II (B) subunit family member... 37.4 0.013
cel:F56D2.6 hypothetical protein; K12820 pre-mRNA-splicing fac... 33.1 0.24
sce:YJL140W RPB4, CTF15; B32; K03012 DNA-directed RNA polymera... 32.7 0.36
dre:286831 lamb4, fa05e06, wu:fa05e06; laminin, beta 4; K06245... 30.8 1.4
hsa:120425 AMICA1, AMICA, CREA7-1, CREA7-4, FLJ37080, Gm638, J... 30.4 1.9
hsa:2 A2M, CPAMD5, DKFZp779B086, S863-7; alpha-2-macroglobulin... 30.0 1.9
mmu:11637 Ak2, Ak-2, D4Ertd220e; adenylate kinase 2 (EC:2.7.4.... 30.0 2.3
xla:379924 ak2, MGC53010; adenylate kinase 2 (EC:2.7.4.3); K00... 28.9 4.7
hsa:204 AK2, ADK2, AK_2; adenylate kinase 2 (EC:2.7.4.3); K009... 28.5 6.8
> tgo:TGME49_031380 DNA-directed RNA polymerase II, putative ;
K03012 DNA-directed RNA polymerase II subunit RPB4
Length=185
Score = 155 bits (393), Expect = 3e-38, Method: Compositional matrix adjust.
Identities = 75/116 (64%), Positives = 93/116 (80%), Gaps = 0/116 (0%)
Query 21 EFKNAKCLNLCELQLILGDQLRLSVARPAEAHNLIKASYEYASRFGKMTVRTAVVEIRKH 80
EF++AKCLNLCELQLIL DQLRL R AEA L+KASY+Y++RF K+ R +V++R +
Sbjct 70 EFRSAKCLNLCELQLILSDQLRLPNTRSAEAQGLLKASYDYSTRFAKLRTRNVIVDLRNN 129
Query 81 LEREGDLQQFELALLVNLLPRTPDEAKAHIPSLIRLSAARLSRIIDTLEVFRVHAS 136
LEREGDL QFE+A LVNLLP++ DEAKA IP+L RL RL RI+D LE+FRVHA+
Sbjct 130 LEREGDLHQFEIAQLVNLLPKSLDEAKALIPTLNRLPGPRLQRILDLLEMFRVHAA 185
> pfa:PFB0245c DNA-directed RNA polymerase II 16 kDa subunit,
putative; K03012 DNA-directed RNA polymerase II subunit RPB4
Length=132
Score = 144 bits (362), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 68/120 (56%), Positives = 94/120 (78%), Gaps = 0/120 (0%)
Query 17 DLGPEFKNAKCLNLCELQLILGDQLRLSVARPAEAHNLIKASYEYASRFGKMTVRTAVVE 76
DLGP+FKN KCLNLCELQLILGDQLRL+ R EA LIK+SY+YA++F + R+++V+
Sbjct 13 DLGPDFKNCKCLNLCELQLILGDQLRLTSKRNEEAQALIKSSYDYANKFAAIKNRSSIVD 72
Query 77 IRKHLEREGDLQQFELALLVNLLPRTPDEAKAHIPSLIRLSAARLSRIIDTLEVFRVHAS 136
IR +LER GDL ++E+A+LVNLLP+T EA+ IPSLIRL+ L+ I++ L ++++ S
Sbjct 73 IRTNLERIGDLHEYEIAMLVNLLPKTILEARYLIPSLIRLNDETLNSILEHLISYKMYVS 132
> bbo:BBOV_III006040 17.m07533; RNA polymerase Rpb4 family protein;
K03012 DNA-directed RNA polymerase II subunit RPB4
Length=129
Score = 131 bits (329), Expect = 7e-31, Method: Compositional matrix adjust.
Identities = 65/119 (54%), Positives = 85/119 (71%), Gaps = 0/119 (0%)
Query 17 DLGPEFKNAKCLNLCELQLILGDQLRLSVARPAEAHNLIKASYEYASRFGKMTVRTAVVE 76
++ E KNAKCLNLCEL LILGDQLRL R A LIK S+EYASRF + R+A+V+
Sbjct 11 EVDEELKNAKCLNLCELHLILGDQLRLQHKRNENAKQLIKTSHEYASRFAILKFRSAIVD 70
Query 77 IRKHLEREGDLQQFELALLVNLLPRTPDEAKAHIPSLIRLSAARLSRIIDTLEVFRVHA 135
IR +EREG L +FE+A +VNLLP++ DEAK+ IPSL R+ R+ I++ LE +R +
Sbjct 71 IRTTIEREGSLHEFEMASVVNLLPKSVDEAKSLIPSLWRIPDERIDDILELLEGYRAQS 129
> tpv:TP02_0359 DNA-directed RNA polymerase II subunit Rpb4; K03012
DNA-directed RNA polymerase II subunit RPB4
Length=104
Score = 95.5 bits (236), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 57/130 (43%), Positives = 76/130 (58%), Gaps = 26/130 (20%)
Query 6 MEASVPESLAGDLGPEFKNAKCLNLCELQLILGDQLRLSVARPAEAHNLIKASYEYASRF 65
ME+ + DL PEFKN+KCLNLCEL LILGDQLRL R
Sbjct 1 MESYNFDPTKFDLDPEFKNSKCLNLCELHLILGDQLRLHHNRND---------------- 44
Query 66 GKMTVRTAVVEIRKHLEREGDLQQFELALLVNLLPRTPDEAKAHIPSLIRLSAARLSRII 125
TAV I ER+G L +FE+A LVNLLP++ DEAK+ IPSL R+ ++++I+
Sbjct 45 ------TAVHTI----ERDGLLHEFEMASLVNLLPKSADEAKSLIPSLCRIPDDKINKIL 94
Query 126 DTLEVFRVHA 135
+ LE +R+ +
Sbjct 95 ELLESYRIQS 104
> cpv:cgd3_4320 hypothetical protein ; K03012 DNA-directed RNA
polymerase II subunit RPB4
Length=124
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 48/95 (50%), Positives = 66/95 (69%), Gaps = 2/95 (2%)
Query 21 EFKNAKCLNLCELQLILGDQLRLSVARPAEAHNLIKASYEYASRFGKMTVRTAVVEIRKH 80
E KCLNL EL+L+L D++R + EAH LIK++Y+Y+ +FGK+ R +V+ IR+
Sbjct 21 ELNGIKCLNLSELRLLLEDRMRTYPSGSDEAHTLIKSAYDYSYKFGKIKNRASVILIREA 80
Query 81 LEREGDLQQFELALLVNLLPRTPDEAKAHI--PSL 113
L+ L +FE+A LVNLLPRTPDEAK + PSL
Sbjct 81 LDETTKLHEFEIASLVNLLPRTPDEAKVSMKHPSL 115
> ath:AT5G09920 NRPB4; NRPB4; DNA-directed RNA polymerase; K03012
DNA-directed RNA polymerase II subunit RPB4
Length=138
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 39/100 (39%), Positives = 55/100 (55%), Gaps = 5/100 (5%)
Query 18 LGPEFKNAKCLNLCELQLILG---DQLRLSVARPA-EAHNLIKASYEYASRFGKMTVRTA 73
+G EF AKCL CE+ LIL +QL+ P + + + S +Y RF + A
Sbjct 14 IGDEFLKAKCLMNCEVSLILEHKFEQLQQISEDPMNQVSQVFEKSLQYVKRFSRYKNPDA 73
Query 74 VVEIRKHLEREGDLQQFELALLVNLLPRTPDEAKAHIPSL 113
V ++R+ L R L +FEL +L NL P T +EA A +PSL
Sbjct 74 VRQVREILSRH-QLTEFELCVLGNLCPETVEEAVAMVPSL 112
> xla:447138 polr2d, MGC85403; polymerase (RNA) II (DNA directed)
polypeptide D; K03012 DNA-directed RNA polymerase II subunit
RPB4
Length=142
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 46/139 (33%), Positives = 66/139 (47%), Gaps = 13/139 (9%)
Query 6 MEASVPESLAGD--------LGP-EFKNAKCLNLCELQLILG--DQLRLSVARPAEAHNL 54
M A E+ +GD L P EF++A+ L E+ ++L Q S E +
Sbjct 1 MAAGGAETHSGDAEEDASQLLFPKEFESAETLLNSEVHMLLEHRKQQNESAEEEQELSEV 60
Query 55 IKASYEYASRFGKMTVRTAVVEIRKHLEREGDLQQFELALLVNLLPRTPDEAKAHIPSLI 114
+ Y +RF + R + +R L + L +FELA L NL P T DEAKA IPSL
Sbjct 61 FMKTLNYTARFSRFKNRETIASVRSLL-LQKKLHRFELACLANLCPETADEAKALIPSLE 119
Query 115 -RLSAARLSRIIDTLEVFR 132
R L +I+D ++ R
Sbjct 120 GRFEEEELQQILDDIQTKR 138
> dre:436588 polr2d, wu:fa95b11, zgc:110435; polymerase (RNA)
II (DNA directed) polypeptide D (EC:2.7.7.6); K03012 DNA-directed
RNA polymerase II subunit RPB4
Length=141
Score = 51.6 bits (122), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 38/115 (33%), Positives = 56/115 (48%), Gaps = 4/115 (3%)
Query 21 EFKNAKCLNLCELQLILG--DQLRLSVARPAEAHNLIKASYEYASRFGKMTVRTAVVEIR 78
EF+NA+ L E+ ++L Q S E + + Y +RF + R + +R
Sbjct 24 EFENAETLLNSEVHMLLEHRKQQNESAEDEQELSEVFMKTLNYTARFSRFKNRETIASVR 83
Query 79 KHLEREGDLQQFELALLVNLLPRTPDEAKAHIPSLI-RLSAARLSRIIDTLEVFR 132
L + L +FELA L NL P +EAKA IPSL R L +I+D ++ R
Sbjct 84 SLL-LQKKLHKFELASLANLCPEAAEEAKALIPSLEGRFEDEELQQILDDIQTKR 137
> mmu:69241 Polr2d, 2310002G05Rik, 2610028L19Rik, HSRBP4, RBP4;
polymerase (RNA) II (DNA directed) polypeptide D (EC:2.7.7.6);
K03012 DNA-directed RNA polymerase II subunit RPB4
Length=142
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 37/115 (32%), Positives = 56/115 (48%), Gaps = 4/115 (3%)
Query 21 EFKNAKCLNLCELQLILG--DQLRLSVARPAEAHNLIKASYEYASRFGKMTVRTAVVEIR 78
EF+ A+ L E+ ++L Q S E + + Y +RF + R + +R
Sbjct 25 EFETAETLLNSEVHMLLEHRKQQNESAEDEQELSEVFMKTLNYTARFSRFKNRETIASVR 84
Query 79 KHLEREGDLQQFELALLVNLLPRTPDEAKAHIPSLI-RLSAARLSRIIDTLEVFR 132
L + L +FELA L NL P T +E+KA IPSL R L +I+D ++ R
Sbjct 85 SLL-LQKKLHKFELACLANLCPETAEESKALIPSLEGRFEDEELQQILDDIQTKR 138
> hsa:5433 POLR2D, HSRBP4, HSRPB4, RBP4, RPB16; polymerase (RNA)
II (DNA directed) polypeptide D (EC:2.7.7.6); K03012 DNA-directed
RNA polymerase II subunit RPB4
Length=142
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 37/115 (32%), Positives = 56/115 (48%), Gaps = 4/115 (3%)
Query 21 EFKNAKCLNLCELQLILG--DQLRLSVARPAEAHNLIKASYEYASRFGKMTVRTAVVEIR 78
EF+ A+ L E+ ++L Q S E + + Y +RF + R + +R
Sbjct 25 EFETAETLLNSEVHMLLEHRKQQNESAEDEQELSEVFMKTLNYTARFSRFKNRETIASVR 84
Query 79 KHLEREGDLQQFELALLVNLLPRTPDEAKAHIPSLI-RLSAARLSRIIDTLEVFR 132
L + L +FELA L NL P T +E+KA IPSL R L +I+D ++ R
Sbjct 85 SLL-LQKKLHKFELACLANLCPETAEESKALIPSLEGRFEDEELQQILDDIQTKR 138
> cel:F43E2.2 rpb-4; RNA Polymerase II (B) subunit family member
(rpb-4); K03012 DNA-directed RNA polymerase II subunit RPB4
Length=144
Score = 37.4 bits (85), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 37/135 (27%), Positives = 61/135 (45%), Gaps = 7/135 (5%)
Query 5 KMEASVPESLAGDLGP-EFKNAKCLNL--CELQLILGDQLRLSVARPA--EAHNLIKASY 59
K + V E A P EF+ C L E+ L+L + + S ++ E + +
Sbjct 9 KSDGQVEEDAAECKFPKEFETTTCDALLTAEVYLLLEHRRQSSESKDEIEEMSEVFIKTL 68
Query 60 EYASRFGKMTVRTAVVEIRKHLEREGDLQQFELALLVNLLPRTPDEAKAHIPSL-IRLSA 118
YA R + R + +R E L +FE+A + NL P +EAKA +PSL ++
Sbjct 69 NYARRMSRFKNRETIRAVRAIF-SEKHLHKFEVAQIANLCPENAEEAKALVPSLENKIEE 127
Query 119 ARLSRIIDTLEVFRV 133
+ L ++ L+ R
Sbjct 128 SELEEVLKDLQSKRT 142
> cel:F56D2.6 hypothetical protein; K12820 pre-mRNA-splicing factor
ATP-dependent RNA helicase DHX15/PRP43 [EC:3.6.4.13]
Length=739
Score = 33.1 bits (74), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 34/69 (49%), Gaps = 4/69 (5%)
Query 38 GDQLRLSVARPAEAHNLIKASYEYASRFGKMTVRTAVVE----IRKHLEREGDLQQFELA 93
D +R ++R + +NL + S ++ SR + +R A+V HLER G +
Sbjct 601 ADTVRTQLSRVMDKYNLRRVSTDFKSRDYYLNIRKALVAGFFMQVAHLERSGHYVTVKDN 660
Query 94 LLVNLLPRT 102
LVNL P T
Sbjct 661 QLVNLHPST 669
> sce:YJL140W RPB4, CTF15; B32; K03012 DNA-directed RNA polymerase
II subunit RPB4
Length=221
Score = 32.7 bits (73), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 26/75 (34%), Positives = 40/75 (53%), Gaps = 2/75 (2%)
Query 55 IKASYEYASRFGKMTVRTAVVEIRKHLEREGDLQQFELALLVNLLPRTPDEAKAHIPSL- 113
+K + +Y + F + + V + + L+ G L FE+A L +L T DEAK IPSL
Sbjct 141 LKNTMQYLTNFSRFRDQETVGAVIQLLKSTG-LHPFEVAQLGSLACDTADEAKTLIPSLN 199
Query 114 IRLSAARLSRIIDTL 128
++S L RI+ L
Sbjct 200 NKISDDELERILKEL 214
> dre:286831 lamb4, fa05e06, wu:fa05e06; laminin, beta 4; K06245
laminin, beta 4
Length=1827
Score = 30.8 bits (68), Expect = 1.4, Method: Composition-based stats.
Identities = 21/83 (25%), Positives = 36/83 (43%), Gaps = 1/83 (1%)
Query 53 NLIKASYEYASRFGKMTVRTAVVEIRKHLEREGDLQQFELALLVNLLPRTPDEAKAHIPS 112
N K YE K + ++ L + D+++ A+L LP++PDE K I
Sbjct 1533 NRTKTKYEQEKTDTKALIEKVKTYLQDELVKPEDIEKLANAVLSIQLPKSPDEIKDMIED 1592
Query 113 LIRLSAARLSRIIDTLEVFRVHA 135
+ ++ A ++ D LE A
Sbjct 1593 IKKI-LANITEFNDDLEYLEKQA 1614
> hsa:120425 AMICA1, AMICA, CREA7-1, CREA7-4, FLJ37080, Gm638,
JAML, MGC118814, MGC118815; adhesion molecule, interacts with
CXADR antigen 1
Length=394
Score = 30.4 bits (67), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 27/48 (56%), Gaps = 4/48 (8%)
Query 72 TAVVEIRKHLEREGDLQQFELALLVNLLPRTPDEAKAHIPSLIRLSAA 119
T + EIR +G+ Q F+ A+++++LP P E H+ LI++
Sbjct 113 TYICEIR----LKGESQVFKKAVVLHVLPEEPKELMVHVGGLIQMGCV 156
> hsa:2 A2M, CPAMD5, DKFZp779B086, S863-7; alpha-2-macroglobulin;
K03910 alpha-2-macroglobulin
Length=1474
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 15/32 (46%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 88 QQFELALLVNLLPRTPDEAKAHIPSLIRLSAA 119
++F AL V LP+T DE KAH I LS +
Sbjct 1337 EEFPFALGVQTLPQTCDEPKAHTSFQISLSVS 1368
> mmu:11637 Ak2, Ak-2, D4Ertd220e; adenylate kinase 2 (EC:2.7.4.3);
K00939 adenylate kinase [EC:2.7.4.3]
Length=239
Score = 30.0 bits (66), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 26/75 (34%), Positives = 34/75 (45%), Gaps = 9/75 (12%)
Query 29 NLCELQLILGDQLRLSVARPAEAHNLIKASYEYASRFGKMTVRTAVVE-IRKHLEREGDL 87
N C L GD LR VA +E +KA+ + GK+ VVE I K+LE
Sbjct 38 NFCVCHLATGDMLRAMVASGSELGKKLKATMDA----GKLVSDEMVVELIEKNLETPSCK 93
Query 88 QQFELALLVNLLPRT 102
F L++ PRT
Sbjct 94 NGF----LLDGFPRT 104
> xla:379924 ak2, MGC53010; adenylate kinase 2 (EC:2.7.4.3); K00939
adenylate kinase [EC:2.7.4.3]
Length=241
Score = 28.9 bits (63), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 36/77 (46%), Gaps = 7/77 (9%)
Query 31 CELQLILGDQLRLSVARPAEAHNLIKASYEYASRFGKMTVRTAVVEIRKHLEREGDLQQF 90
C L GD LR VA +E +KA+ + GK+ VVE+ +E+ D
Sbjct 42 CVCHLATGDMLRAMVASGSELGKRLKATMDA----GKLVSDEMVVEL---IEKNLDTPPC 94
Query 91 ELALLVNLLPRTPDEAK 107
+ L++ PRT +A+
Sbjct 95 KKGFLLDGFPRTVKQAE 111
> hsa:204 AK2, ADK2, AK_2; adenylate kinase 2 (EC:2.7.4.3); K00939
adenylate kinase [EC:2.7.4.3]
Length=224
Score = 28.5 bits (62), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 21/55 (38%), Positives = 27/55 (49%), Gaps = 5/55 (9%)
Query 29 NLCELQLILGDQLRLSVARPAEAHNLIKASYEYASRFGKMTVRTAVVE-IRKHLE 82
N C L GD LR VA +E +KA+ + GK+ VVE I K+LE
Sbjct 38 NFCVCHLATGDMLRAMVASGSELGKKLKATMDA----GKLVSDEMVVELIEKNLE 88
Lambda K H
0.321 0.135 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2362384368
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40