bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1834_orf5
Length=267
Score E
Sequences producing significant alignments: (Bits) Value
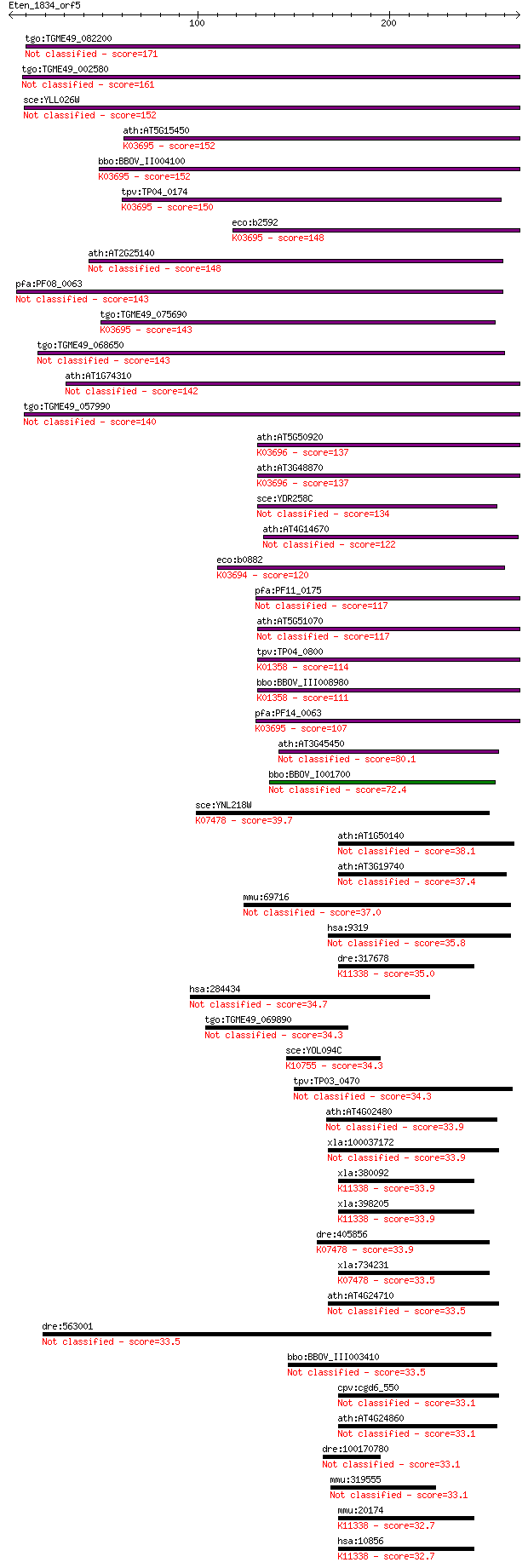
tgo:TGME49_082200 clpB protein, putative 171 4e-42
tgo:TGME49_002580 heat shock protein, putative (EC:3.4.21.53) 161 2e-39
sce:YLL026W HSP104; Heat shock protein that cooperates with Yd... 152 1e-36
ath:AT5G15450 CLPB3; CLPB3 (CASEIN LYTIC PROTEINASE B3); ATP b... 152 1e-36
bbo:BBOV_II004100 18.m06340; ClpB; K03695 ATP-dependent Clp pr... 152 1e-36
tpv:TP04_0174 hypothetical protein; K03695 ATP-dependent Clp p... 150 4e-36
eco:b2592 clpB, ECK2590, htpM, JW2573; protein disaggregation ... 148 2e-35
ath:AT2G25140 CLPB4; CLPB4 (CASEIN LYTIC PROTEINASE B4); ATP b... 148 3e-35
pfa:PF08_0063 ClpB protein, putative 143 5e-34
tgo:TGME49_075690 chaperone clpB 1 protein, putative (EC:3.4.2... 143 6e-34
tgo:TGME49_068650 clp ATP-binding chain B1, putative (EC:3.4.2... 143 7e-34
ath:AT1G74310 ATHSP101 (ARABIDOPSIS THALIANA HEAT SHOCK PROTEI... 142 1e-33
tgo:TGME49_057990 heat shock protein, putative (EC:3.4.21.53) 140 6e-33
ath:AT5G50920 CLPC1; CLPC1; ATP binding / ATP-dependent peptid... 137 3e-32
ath:AT3G48870 HSP93-III; ATP binding / ATPase/ DNA binding / n... 137 4e-32
sce:YDR258C HSP78; Hsp78p 134 3e-31
ath:AT4G14670 CLPB2; CLPB2; ATP binding / nucleoside-triphosph... 122 1e-27
eco:b0882 clpA, ECK0873, JW0866, lopD; ATPase and specificity ... 120 5e-27
pfa:PF11_0175 heat shock protein 101, putative 117 4e-26
ath:AT5G51070 ERD1; ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1); ... 117 5e-26
tpv:TP04_0800 ATP-dependent Clp protease ATP-binding subunit (... 114 5e-25
bbo:BBOV_III008980 17.m07783; Clp amino terminal domain contai... 111 2e-24
pfa:PF14_0063 ATP-dependent CLP protease, putative; K03695 ATP... 107 3e-23
ath:AT3G45450 Clp amino terminal domain-containing protein 80.1 7e-15
bbo:BBOV_I001700 19.m02115; chaperone clpB 72.4 2e-12
sce:YNL218W MGS1; Mgs1p; K07478 putative ATPase 39.7 0.011
ath:AT1G50140 ATP binding / ATPase/ nucleoside-triphosphatase/... 38.1 0.038
ath:AT3G19740 ATP binding / ATPase/ nucleoside-triphosphatase/... 37.4 0.062
mmu:69716 Trip13, 2410002G23Rik, D13Ertd328e; thyroid hormone ... 37.0 0.074
hsa:9319 TRIP13, 16E1BP; thyroid hormone receptor interactor 13 35.8
dre:317678 ruvbl2, reptin, wu:fi25f01, zreptin; RuvB-like 2 (E... 35.0 0.28
hsa:284434 NWD1, MGC134940; NACHT and WD repeat domain contain... 34.7 0.35
tgo:TGME49_069890 M16 family peptidase, putative (EC:3.4.24.56) 34.3 0.46
sce:YOL094C RFC4; Rfc4p; K10755 replication factor C subunit 2/4 34.3
tpv:TP03_0470 origin recognition complex 1 protein 34.3 0.54
ath:AT4G02480 AAA-type ATPase family protein 33.9 0.57
xla:100037172 trip13; thyroid hormone receptor interactor 13 33.9
xla:380092 ruvbl2, MGC52995; RuvB-like protein 2; K11338 RuvB-... 33.9 0.59
xla:398205 ruvbl2, tip48, xReptin; RuvB-like 2 (EC:3.6.4.12); ... 33.9 0.60
dre:405856 MGC85976; zgc:85976; K07478 putative ATPase 33.9 0.68
xla:734231 wrnip1; Werner helicase interacting protein 1; K074... 33.5 0.74
ath:AT4G24710 ATP binding / ATPase/ nucleoside-triphosphatase/... 33.5 0.84
dre:563001 hypothetical LOC563001 33.5 0.86
bbo:BBOV_III003410 17.m10506; hypothetical protein 33.5 0.90
cpv:cgd6_550 Pch2p like AAA ATpase 33.1 1.0
ath:AT4G24860 AAA-type ATPase family protein 33.1 1.0
dre:100170780 zgc:195077 33.1 1.1
mmu:319555 Nwd1, A230063L24Rik; NACHT and WD repeat domain con... 33.1 1.2
mmu:20174 Ruvbl2, MGC144733, MGC144734, mp47, p47; RuvB-like p... 32.7 1.3
hsa:10856 RUVBL2, ECP51, INO80J, REPTIN, RVB2, TIH2, TIP48, TI... 32.7 1.3
> tgo:TGME49_082200 clpB protein, putative
Length=970
Score = 171 bits (432), Expect = 4e-42, Method: Compositional matrix adjust.
Identities = 104/270 (38%), Positives = 161/270 (59%), Gaps = 13/270 (4%)
Query 10 IEKCQGDYDFIRHSVRDFIVGLPRSSVP--INLGESGPSVALRNALRKAWKMAEFEGAPI 67
+E+C + ++ ++ + ++ LP S+ ++ ES P + R ALR + + I
Sbjct 105 LEQCGANVYELKKAMSNVLIKLPTSADVEIVDELESTPQLK-RVALRALEIAQKDKKQTI 163
Query 68 GVQHLYLALVEILAFQKIMVKSGCDLRLFKHEIVEQFKGR-QRGVAPTSFQEMVGD---- 122
G++HL +L E +K + ++GC++++F ++ + ++ VA S ++ +
Sbjct 164 GMEHLITSLFEENNLRKSLEQAGCNVKVFLEHSTTMYENKFKQDVASRSMKKQPTEGSPA 223
Query 123 ----KQGTSDA-FVSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQILSRKMTKVPMLVGE 177
K SD FV +GVD++ +A +G L PVVGR++EI + +LSRK P LVGE
Sbjct 224 TSRAKPDASDEEFVQSFGVDMTKLAAEGKLEPVVGRNKEIKEVLTVLSRKGKGNPCLVGE 283
Query 178 PGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQ 237
PGVGKTAV+EGLAQR+VEG VP+SL + +FA+DL +L AG+ RGEFEKRMK ++ Y
Sbjct 284 PGVGKTAVVEGLAQRLVEGMVPKSLENKILFAVDLGALIAGATYRGEFEKRMKALIRYAV 343
Query 238 NNVEEVILFVDEIHTLIGAAGGVCRNDRAN 267
N VILF+DE+H L+GA D AN
Sbjct 344 NQEGRVILFIDELHMLMGAGKSDGTMDAAN 373
> tgo:TGME49_002580 heat shock protein, putative (EC:3.4.21.53)
Length=983
Score = 161 bits (408), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 94/262 (35%), Positives = 149/262 (56%), Gaps = 8/262 (3%)
Query 8 AVIEKCQGDYDFIRHSVRDFIVGLPRSSVPINLGESGPSVALRNALRKAWKMAEFEGAP- 66
+EK G +R + + ++ +P+S P++ GE+ L+ + A+ A+ G
Sbjct 143 TALEKAGGKSADLRKDMHNLLIRMPQSVTPLSYGEARSRSELKRVIIDAYNEAQSRGHRL 202
Query 67 IGVQHLYLALVEILAFQKIMVKSGCDLRLFKHEIVEQFKGRQRGVAPTSFQEMVGDKQGT 126
+GV L AL F+ + G D+ +F + F + T +E T
Sbjct 203 VGVHDLVTALARCDRFRARLANVGTDVSVFLKLVTSAF------IVHTEAEERKATPART 256
Query 127 -SDAFVSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQILSRKMTKVPMLVGEPGVGKTAV 185
S ++ +GVD++ A++G + V GR+ EI++I ++SR +L+GEPGVGKTAV
Sbjct 257 GSQEWIKTFGVDMTEQAKEGKIGTVTGREAEIEQITSVMSRMSKANCLLIGEPGVGKTAV 316
Query 186 IEGLAQRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQNNVEEVIL 245
+EGLA+RIV+G VP++L ++F+LD+ SL +GS+MRGEFE+RMK ++ YL + IL
Sbjct 317 VEGLAKRIVDGDVPDALLGVQVFSLDVGSLLSGSSMRGEFERRMKGILDYLFASDRSTIL 376
Query 246 FVDEIHTLIGAAGGVCRNDRAN 267
F+DEIHTL+GA D AN
Sbjct 377 FIDEIHTLMGAGKADGPMDAAN 398
> sce:YLL026W HSP104; Heat shock protein that cooperates with
Ydj1p (Hsp40) and Ssa1p (Hsp70) to refold and reactivate previously
denatured, aggregated proteins; responsive to stresses
including: heat, ethanol, and sodium arsenite; involved in
[PSI+] propagation
Length=908
Score = 152 bits (385), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 91/261 (34%), Positives = 152/261 (58%), Gaps = 17/261 (6%)
Query 9 VIEKCQGDYDFIRHSVRDFIVGLPRSS-VPINLGESGPSVALRNALRKAWKM-AEFEGAP 66
+IEK + DYD + V +V +P+ P E PS AL L+ A K+ + + +
Sbjct 54 LIEKGRYDYDLFKKVVNRNLVRIPQQQPAP---AEITPSYALGKVLQDAAKIQKQQKDSF 110
Query 67 IGVQHLYLALVEILAFQKIMVKSGCDLRLFKHEIVEQFKGRQRGVAPTSFQEMVGDKQGT 126
I H+ AL + Q+I ++ D+ K + +E +G R + G T
Sbjct 111 IAQDHILFALFNDSSIQQIFKEAQVDIEAIKQQALE-LRGNTRI-------DSRGADTNT 162
Query 127 SDAFVSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQILSRKMTKVPMLVGEPGVGKTAVI 186
++S+Y +D++ A+QG L PV+GR+EEI ++L+R++ P L+GEPG+GKTA+I
Sbjct 163 PLEYLSKYAIDMTEQARQGKLDPVIGREEEIRSTIRVLARRIKSNPCLIGEPGIGKTAII 222
Query 187 EGLAQRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQNNVEEVILF 246
EG+AQRI++ VP L+ ++F+LDL +L+AG+ +G+FE+R K V+ ++ + ++LF
Sbjct 223 EGVAQRIIDDDVPTILQGAKLFSLDLAALTAGAKYKGDFEERFKGVLKEIEESKTLIVLF 282
Query 247 VDEIHTLIGAAGGVCRNDRAN 267
+DEIH L+G ++D AN
Sbjct 283 IDEIHMLMGNG----KDDAAN 299
> ath:AT5G15450 CLPB3; CLPB3 (CASEIN LYTIC PROTEINASE B3); ATP
binding / ATPase/ nucleoside-triphosphatase/ nucleotide binding
/ protein binding; K03695 ATP-dependent Clp protease ATP-binding
subunit ClpB
Length=968
Score = 152 bits (384), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 85/210 (40%), Positives = 132/210 (62%), Gaps = 15/210 (7%)
Query 61 EFEGAPIGVQHLYLALVEILAFQKIMVKSGCDLRLFKHEI---VEQFKGRQRGVAPTSFQ 117
+ + + + V+HL LA + F K + K D ++ + + +E +G+Q +
Sbjct 176 DLKDSYVSVEHLVLAFADDKRFGKQLFK---DFQISERSLKSAIESIRGKQSVIDQ---- 228
Query 118 EMVGDKQGTSDAFVSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQILSRKMTKVPMLVGE 177
D +G +A + +YG DL+ MA++G L PV+GRD+EI R QILSR+ P+L+GE
Sbjct 229 ----DPEGKYEA-LEKYGKDLTAMAREGKLDPVIGRDDEIRRCIQILSRRTKNNPVLIGE 283
Query 178 PGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQ 237
PGVGKTA+ EGLAQRIV+G VP++L R++ +LD+ +L AG+ RGEFE R+K V+ +
Sbjct 284 PGVGKTAISEGLAQRIVQGDVPQALMNRKLISLDMGALIAGAKYRGEFEDRLKAVLKEVT 343
Query 238 NNVEEVILFVDEIHTLIGAAGGVCRNDRAN 267
++ ++ILF+DEIHT++GA D N
Sbjct 344 DSEGQIILFIDEIHTVVGAGATNGAMDAGN 373
> bbo:BBOV_II004100 18.m06340; ClpB; K03695 ATP-dependent Clp
protease ATP-binding subunit ClpB
Length=931
Score = 152 bits (383), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 92/230 (40%), Positives = 133/230 (57%), Gaps = 27/230 (11%)
Query 48 ALRNALRKAWKM-AEFEGAPIGVQHLYLALV-EILAFQKIMVKSGCDLRLFKHEI----- 100
L+N L ++ +E+ I V+HL LAL E F K L KH++
Sbjct 127 TLQNVLTVTRRIKSEYNDHFISVEHLLLALACEDTKFTKPW--------LTKHKVGYDKL 178
Query 101 ---VEQFKGRQRGVAPTSFQEMVGDKQGTSDAFVSQYGVDLSFMAQQGLLPPVVGRDEEI 157
VE +G+ R V + + + G + +Y DL+ MA+ G L PV+GRD EI
Sbjct 179 KRAVESVRGK-RKVTSKNPEMLFG--------VLEKYSKDLTMMARSGKLDPVIGRDNEI 229
Query 158 DRIAQILSRKMTKVPMLVGEPGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSA 217
R +ILSR+ P+L+G+PGVGKTA+ EGLA RIV G VP+SL+ R+ +LDL S+ A
Sbjct 230 RRTVEILSRRTKNNPILLGDPGVGKTAIAEGLANRIVSGDVPDSLKNTRVISLDLASMLA 289
Query 218 GSAMRGEFEKRMKEVVSYLQNNVEEVILFVDEIHTLIGAAGGVCRNDRAN 267
GS RGEFE+R+K ++ +Q++ E+I+F+DEIHT++GA D N
Sbjct 290 GSQYRGEFEERLKNILKEVQDSQGEIIMFIDEIHTVVGAGDAQGAMDAGN 339
> tpv:TP04_0174 hypothetical protein; K03695 ATP-dependent Clp
protease ATP-binding subunit ClpB
Length=985
Score = 150 bits (379), Expect = 4e-36, Method: Composition-based stats.
Identities = 78/200 (39%), Positives = 125/200 (62%), Gaps = 12/200 (6%)
Query 60 AEFEGAPIGVQHLYLALV--EILAFQKIMVKSGCDLRLFKHEIVEQFKGRQRGVAPTSFQ 117
+EF I V HL L L + F+ + ++ L K ++ +G+++
Sbjct 200 SEFGDKYISVDHLLLGLAAEDTKFFRPYLTRNKVTLEKLKDSVL-SIRGKRK-------- 250
Query 118 EMVGDKQGTSDAFVSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQILSRKMTKVPMLVGE 177
+ S ++++ DL+ MA+ G L PV+GRD EI R +ILSR+ P+L+G+
Sbjct 251 -ITSRNTENSYKLLNKFSKDLTDMARNGKLDPVIGRDNEIRRTIEILSRRTKNNPVLLGD 309
Query 178 PGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQ 237
PGVGKTA+ EGLA RIV G VP+SL+ R++ +LD+ ++ AG+ RGEFE+R+KE++S ++
Sbjct 310 PGVGKTAIAEGLANRIVSGDVPDSLKNRKVLSLDIAAIVAGTMYRGEFEERLKEILSEIE 369
Query 238 NNVEEVILFVDEIHTLIGAA 257
N+ E+++F+DEIHTL+GA
Sbjct 370 NSQGEIVMFIDEIHTLVGAG 389
> eco:b2592 clpB, ECK2590, htpM, JW2573; protein disaggregation
chaperone; K03695 ATP-dependent Clp protease ATP-binding subunit
ClpB
Length=857
Score = 148 bits (374), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 77/152 (50%), Positives = 103/152 (67%), Gaps = 3/152 (1%)
Query 118 EMVGDKQGTSDA--FVSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQILSRKMTKVPMLV 175
E V D QG D + +Y +DL+ A+QG L PV+GRDEEI R Q+L R+ P+L+
Sbjct 147 ESVND-QGAEDQRQALKKYTIDLTERAEQGKLDPVIGRDEEIRRTIQVLQRRTKNNPVLI 205
Query 176 GEPGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSY 235
GEPGVGKTA++EGLAQRI+ G VPE L+ RR+ ALD+ +L AG+ RGEFE+R+K V++
Sbjct 206 GEPGVGKTAIVEGLAQRIINGEVPEGLKGRRVLALDMGALVAGAKYRGEFEERLKGVLND 265
Query 236 LQNNVEEVILFVDEIHTLIGAAGGVCRNDRAN 267
L VILF+DE+HT++GA D N
Sbjct 266 LAKQEGNVILFIDELHTMVGAGKADGAMDAGN 297
> ath:AT2G25140 CLPB4; CLPB4 (CASEIN LYTIC PROTEINASE B4); ATP
binding / ATPase/ nucleoside-triphosphatase/ nucleotide binding
/ protein binding
Length=964
Score = 148 bits (373), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 89/218 (40%), Positives = 129/218 (59%), Gaps = 16/218 (7%)
Query 43 SGPSVALRNALRKAWKMAEFEGAPIGVQHLYLALVEILAF-QKIMVKSGCDLRLFKHEIV 101
S SV L NA R M + + + V+H LA F Q+ D+++ K + +
Sbjct 166 SSLSVILENAKRHKKDMLD---SYVSVEHFLLAYYSDTRFGQEFFRDMKLDIQVLK-DAI 221
Query 102 EQFKGRQRGVAPTSFQEMVGDKQGTSD-AFVSQYGVDLSFMAQQGLLPPVVGRDEEIDRI 160
+ +G QR V D+ S + +YG DL+ MA++G L PV+GRD+EI R
Sbjct 222 KDVRGDQR----------VTDRNPESKYQALEKYGNDLTEMARRGKLDPVIGRDDEIRRC 271
Query 161 AQILSRKMTKVPMLVGEPGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSAGSA 220
QIL R+ P+++GEPGVGKTA+ EGLAQRIV G VPE L R++ +LD+ SL AG+
Sbjct 272 IQILCRRTKNNPVIIGEPGVGKTAIAEGLAQRIVRGDVPEPLMNRKLISLDMGSLLAGAK 331
Query 221 MRGEFEKRMKEVVSYLQNNVEEVILFVDEIHTLIGAAG 258
RG+FE+R+K V+ + + + ILF+DEIHT++GA
Sbjct 332 FRGDFEERLKAVMKEVSASNGQTILFIDEIHTVVGAGA 369
> pfa:PF08_0063 ClpB protein, putative
Length=1070
Score = 143 bits (361), Expect = 5e-34, Method: Composition-based stats.
Identities = 89/258 (34%), Positives = 155/258 (60%), Gaps = 16/258 (6%)
Query 5 ISRAVIEKCQGDYDFIRHSVRDFIVGLPRSSVPINLGESGP-SVALRNALRKAWKMA-EF 62
++ ++++ D + + D++ P+ +P GE L+ L + ++ EF
Sbjct 196 LAERILKESGIDTQLLVQEIDDYLKKQPK--MPSGFGEQKILGRTLQTVLSTSKRLKKEF 253
Query 63 EGAPIGVQHLYLALV-EILAFQK-IMVKSGCDLRLFKHEIVEQFKGRQRGVAPTSFQEMV 120
I ++HL L+++ E F + ++K + K + VE+ +G+++ + T EM
Sbjct 254 NDEYISIEHLLLSIISEDSKFTRPWLLKYNVNYEKVK-KAVEKIRGKKKVTSKTP--EM- 309
Query 121 GDKQGTSDAFVSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQILSRKMTKVPMLVGEPGV 180
T A + +Y DL+ +A+ G L PV+GRD EI R QILSR+ P+L+G+PGV
Sbjct 310 -----TYQA-LEKYSRDLTALARAGKLDPVIGRDNEIRRAIQILSRRTKNNPILLGDPGV 363
Query 181 GKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQNNV 240
GKTA++EGLA +IV+G VP+SL+ R++ +LD+ SL AG+ RG+FE+R+K ++ +Q+
Sbjct 364 GKTAIVEGLAIKIVQGDVPDSLKGRKLVSLDMSSLIAGAKYRGDFEERLKSILKEVQDAE 423
Query 241 EEVILFVDEIHTLIGAAG 258
+V++F+DEIHT++GA
Sbjct 424 GQVVMFIDEIHTVVGAGA 441
> tgo:TGME49_075690 chaperone clpB 1 protein, putative (EC:3.4.21.53);
K03695 ATP-dependent Clp protease ATP-binding subunit
ClpB
Length=898
Score = 143 bits (361), Expect = 6e-34, Method: Compositional matrix adjust.
Identities = 84/212 (39%), Positives = 131/212 (61%), Gaps = 19/212 (8%)
Query 49 LRNALRKAWKMA-EFEGAPIGVQHLYLALV-EILAFQKIMVKSGCDLRLFK-HEIVEQFK 105
L+N L A + EF+ + V+HL LAL E F + + G ++ K VE +
Sbjct 233 LQNVLTAAQRYKREFKDQYLSVEHLVLALAAEDTKFTRPFLTRG-NVSFNKLRSAVEDIR 291
Query 106 GRQRGVAPT---SFQEMVGDKQGTSDAFVSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQ 162
G+++ + ++Q + +Y DL+ A+ G L PV+GRD+EI R Q
Sbjct 292 GKKKVTSKNPELAYQAL------------ERYSRDLTAAARAGKLDPVIGRDDEIRRTIQ 339
Query 163 ILSRKMTKVPMLVGEPGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSAGSAMR 222
ILSR+ P+L+G+PGVGKTA++EGLAQRI+ G VP+SL+ RR+ +LD+ +L AG+ R
Sbjct 340 ILSRRTKNNPVLLGDPGVGKTAIVEGLAQRIISGDVPDSLKGRRVISLDMAALIAGAKYR 399
Query 223 GEFEKRMKEVVSYLQNNVEEVILFVDEIHTLI 254
GEFE+R+K V+ +Q+ +V++F+DEIHT++
Sbjct 400 GEFEERLKAVLKEVQDAEGDVVMFIDEIHTVV 431
> tgo:TGME49_068650 clp ATP-binding chain B1, putative (EC:3.4.21.53)
Length=929
Score = 143 bits (360), Expect = 7e-34, Method: Compositional matrix adjust.
Identities = 95/257 (36%), Positives = 146/257 (56%), Gaps = 36/257 (14%)
Query 16 DYDFIRHSVRDFIVGLPRSSVPINLGES-GPSV----ALRNALRKAWKMAEFEGAPIGVQ 70
D D +R V DF+ P +V N + GP + + N+LR W E+ I V+
Sbjct 73 DLDKLRSGVVDFVERHP--TVSDNSTRTLGPVLQKVLSSANSLRLQWH-DEY----ISVE 125
Query 71 HLYLALVE-----ILAFQKIMVKSGCDLRLFKHEIVEQFKGRQRGVAPT---SFQEMVGD 122
HL AL + ++ F K + D+R + ++ +G +R T S+Q +
Sbjct 126 HLAAALADEDSRFLVRFLKESKLTANDIR----QAIKAIRGTRRVNTKTPEVSYQSL--- 178
Query 123 KQGTSDAFVSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQILSRKMTKVPMLVGEPGVGK 182
+YG DL+ A L PV+GRD+E+ R+ QILSR+ P+++G+PGVGK
Sbjct 179 ---------KKYGRDLTEAAMANELDPVIGRDKEVRRVIQILSRRTKNNPIILGDPGVGK 229
Query 183 TAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQNNVEE 242
TA+ EGLAQRIV G VP++L R++ +LDL +L AG+ +RGEFE+R+K V+ +Q + +
Sbjct 230 TAIAEGLAQRIVSGDVPDTLAGRQLISLDLGALLAGAKLRGEFEERLKSVIREVQESSGQ 289
Query 243 VILFVDEIHTLIGAAGG 259
+ILF+DEIH ++GA
Sbjct 290 IILFIDEIHMVVGAGSA 306
> ath:AT1G74310 ATHSP101 (ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN
101); ATP binding / ATPase/ nucleoside-triphosphatase/ nucleotide
binding / protein binding
Length=911
Score = 142 bits (358), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 94/238 (39%), Positives = 138/238 (57%), Gaps = 13/238 (5%)
Query 31 LPRSSVPINLGESGPSVALRNALRKAWKMAEFEG-APIGVQHLYLALVEILAFQKIMVKS 89
LP S P + + S +L +R+A + G + V L + L+E + ++ +
Sbjct 73 LPSQSPPPD--DIPASSSLIKVIRRAQAAQKSRGDTHLAVDQLIMGLLEDSQIRDLLNEV 130
Query 90 GCDLRLFKHEIVEQFKGRQRGVAPTSFQEMVGDKQGTSDAFVSQYGVDLSFMAQQGLLPP 149
G K E VE+ +G++ + GD T+ + YG DL + Q G L P
Sbjct 131 GVATARVKSE-VEKLRGKE----GKKVESASGD---TNFQALKTYGRDL--VEQAGKLDP 180
Query 150 VVGRDEEIDRIAQILSRKMTKVPMLVGEPGVGKTAVIEGLAQRIVEGAVPESLRKRRIFA 209
V+GRDEEI R+ +ILSR+ P+L+GEPGVGKTAV+EGLAQRIV+G VP SL R+ +
Sbjct 181 VIGRDEEIRRVVRILSRRTKNNPVLIGEPGVGKTAVVEGLAQRIVKGDVPNSLTDVRLIS 240
Query 210 LDLLSLSAGSAMRGEFEKRMKEVVSYLQNNVEEVILFVDEIHTLIGAAGGVCRNDRAN 267
LD+ +L AG+ RGEFE+R+K V+ +++ +VILF+DEIH ++GA D AN
Sbjct 241 LDMGALVAGAKYRGEFEERLKSVLKEVEDAEGKVILFIDEIHLVLGAGKTEGSMDAAN 298
> tgo:TGME49_057990 heat shock protein, putative (EC:3.4.21.53)
Length=921
Score = 140 bits (352), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 89/260 (34%), Positives = 148/260 (56%), Gaps = 14/260 (5%)
Query 9 VIEKCQGDYDFIRHSVRDFIVGLPRSSVPINLGESGPSVALRNALRKAWKM-AEFEGAPI 67
V+ +C GD++ ++ V ++ P+ P + P+ +L LR A + + + +
Sbjct 49 VLSQCPGDFEQLKEDVHLAVLKFPQQHPPPDF--PSPNHSLMAVLRHAKDIQKKMNDSLM 106
Query 68 GVQHLYLALVEILAFQKIMVKSGCDLRLFKHEIVEQFKGRQRGVAPTSFQEMVGDKQGTS 127
L+ ALV+ + + +G F + +E+ RG S + D
Sbjct 107 SADSLFSALVQEKGIRSHLTAAG-----FMMKQIEEKAKSVRG----SKKIASSDDDANF 157
Query 128 DAFVSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQILSRKMTKVPMLVGEPGVGKTAVIE 187
+A + +YG D + +A++G L PV+GR++EI R+ +IL R+ P+L+GEPGVGK+AV+E
Sbjct 158 EA-LKKYGTDFTDLAEKGKLDPVIGREDEIRRVIRILCRRTKNNPVLIGEPGVGKSAVVE 216
Query 188 GLAQRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQNNVEEVILFV 247
GLA+RIVE VP +LR R+ +LD+ SL AG+ RGEFE+R+ V+ +++ ++ILF+
Sbjct 217 GLARRIVEHDVPSNLRC-RLVSLDVGSLIAGAKFRGEFEERLTAVLQEVKDAAGKIILFI 275
Query 248 DEIHTLIGAAGGVCRNDRAN 267
DEIH ++GA D AN
Sbjct 276 DEIHVILGAGKTEGALDAAN 295
> ath:AT5G50920 CLPC1; CLPC1; ATP binding / ATP-dependent peptidase/
ATPase; K03696 ATP-dependent Clp protease ATP-binding
subunit ClpC
Length=929
Score = 137 bits (346), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 66/137 (48%), Positives = 98/137 (71%), Gaps = 1/137 (0%)
Query 131 VSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQILSRKMTKVPMLVGEPGVGKTAVIEGLA 190
+ +YG +L+ +A++G L PVVGR +I+R+ QIL R+ P L+GEPGVGKTA+ EGLA
Sbjct 257 LEEYGTNLTKLAEEGKLDPVVGRQPQIERVVQILGRRTKNNPCLIGEPGVGKTAIAEGLA 316
Query 191 QRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQNNVEEVILFVDEI 250
QRI G VPE++ +++ LD+ L AG+ RGEFE+R+K+++ ++ + +E+ILF+DE+
Sbjct 317 QRIASGDVPETIEGKKVITLDMGLLVAGTKYRGEFEERLKKLMEEIRQS-DEIILFIDEV 375
Query 251 HTLIGAAGGVCRNDRAN 267
HTLIGA D AN
Sbjct 376 HTLIGAGAAEGAIDAAN 392
> ath:AT3G48870 HSP93-III; ATP binding / ATPase/ DNA binding /
nuclease/ nucleoside-triphosphatase/ nucleotide binding / protein
binding; K03696 ATP-dependent Clp protease ATP-binding
subunit ClpC
Length=952
Score = 137 bits (345), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 66/137 (48%), Positives = 98/137 (71%), Gaps = 1/137 (0%)
Query 131 VSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQILSRKMTKVPMLVGEPGVGKTAVIEGLA 190
+ +YG +L+ +A++G L PVVGR +I+R+ QIL+R+ P L+GEPGVGKTA+ EGLA
Sbjct 278 LEEYGTNLTKLAEEGKLDPVVGRQPQIERVVQILARRTKNNPCLIGEPGVGKTAIAEGLA 337
Query 191 QRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQNNVEEVILFVDEI 250
QRI G VPE++ + + LD+ L AG+ RGEFE+R+K+++ ++ + +E+ILF+DE+
Sbjct 338 QRIASGDVPETIEGKTVITLDMGLLVAGTKYRGEFEERLKKLMEEIRQS-DEIILFIDEV 396
Query 251 HTLIGAAGGVCRNDRAN 267
HTLIGA D AN
Sbjct 397 HTLIGAGAAEGAIDAAN 413
> sce:YDR258C HSP78; Hsp78p
Length=811
Score = 134 bits (338), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 66/125 (52%), Positives = 92/125 (73%), Gaps = 0/125 (0%)
Query 131 VSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQILSRKMTKVPMLVGEPGVGKTAVIEGLA 190
+ Q+G +L+ +A+ G L PV+GRDEEI R QILSR+ P L+G GVGKTA+I+GLA
Sbjct 98 LEQFGTNLTKLARDGKLDPVIGRDEEIARAIQILSRRTKNNPCLIGRAGVGKTALIDGLA 157
Query 191 QRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQNNVEEVILFVDEI 250
QRIV G VP+SL+ + + ALDL SL AG+ RGEFE+R+K+V+ + +VI+F+DE+
Sbjct 158 QRIVAGEVPDSLKDKDLVALDLGSLIAGAKYRGEFEERLKKVLEEIDKANGKVIVFIDEV 217
Query 251 HTLIG 255
H L+G
Sbjct 218 HMLLG 222
> ath:AT4G14670 CLPB2; CLPB2; ATP binding / nucleoside-triphosphatase/
nucleotide binding / protein binding
Length=623
Score = 122 bits (306), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 61/133 (45%), Positives = 91/133 (68%), Gaps = 2/133 (1%)
Query 134 YGVDLSFMAQQGLLPPVVGRDEEIDRIAQILSRKMTKVPMLVGEPGVGKTAVIEGLAQRI 193
YG DL + Q G L PV+GR EI R+ ++LSR+ P+L+GEPGVGKTAV+EGLAQRI
Sbjct 132 YGTDL--VEQAGKLDPVIGRHREIRRVIEVLSRRTKNNPVLIGEPGVGKTAVVEGLAQRI 189
Query 194 VEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQNNVEEVILFVDEIHTL 253
++G VP +L ++ +L+ ++ AG+ +RG+FE+R+K V+ ++ +V+LF+DEIH
Sbjct 190 LKGDVPINLTGVKLISLEFGAMVAGTTLRGQFEERLKSVLKAVEEAQGKVVLFIDEIHMA 249
Query 254 IGAAGGVCRNDRA 266
+GA D A
Sbjct 250 LGACKASGSTDAA 262
> eco:b0882 clpA, ECK0873, JW0866, lopD; ATPase and specificity
subunit of ClpA-ClpP ATP-dependent serine protease, chaperone
activity; K03694 ATP-dependent Clp protease ATP-binding
subunit ClpA
Length=758
Score = 120 bits (301), Expect = 5e-27, Method: Compositional matrix adjust.
Identities = 63/150 (42%), Positives = 98/150 (65%), Gaps = 7/150 (4%)
Query 110 GVAPTSFQEMVGDKQGTSDAFVSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQILSRKMT 169
G P S ++ G+++ + + +L+ +A+ G + P++GR++E++R Q+L R+
Sbjct 154 GSQPNSEEQAGGEER------MENFTTNLNQLARVGGIDPLIGREKELERAIQVLCRRRK 207
Query 170 KVPMLVGEPGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRM 229
P+LVGE GVGKTA+ EGLA RIV+G VPE + I++LD+ SL AG+ RG+FEKR
Sbjct 208 NNPLLVGESGVGKTAIAEGLAWRIVQGDVPEVMADCTIYSLDIGSLLAGTKYRGDFEKRF 267
Query 230 KEVVSYLQNNVEEVILFVDEIHTLIGAAGG 259
K ++ L+ + ILF+DEIHT+IGA
Sbjct 268 KALLKQLEQDTNS-ILFIDEIHTIIGAGAA 296
> pfa:PF11_0175 heat shock protein 101, putative
Length=906
Score = 117 bits (294), Expect = 4e-26, Method: Composition-based stats.
Identities = 57/138 (41%), Positives = 85/138 (61%), Gaps = 0/138 (0%)
Query 130 FVSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQILSRKMTKVPMLVGEPGVGKTAVIEGL 189
++ Q+G +++ + G L + GRDEEI I + L R P+LVG PG GKT ++EGL
Sbjct 189 YIEQFGSNMNEKVRNGKLQGIYGRDEEIRAIIESLLRYNKNSPVLVGNPGTGKTTIVEGL 248
Query 190 AQRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQNNVEEVILFVDE 249
RI +G VP+ L+ + +L+ ++G++ RGEFE RMK ++ L+N ++ILFVDE
Sbjct 249 VYRIEKGDVPKELQGYTVISLNFRKFTSGTSYRGEFETRMKNIIKELKNKKNKIILFVDE 308
Query 250 IHTLIGAAGGVCRNDRAN 267
IH L+GA D AN
Sbjct 309 IHLLLGAGKAEGGTDAAN 326
> ath:AT5G51070 ERD1; ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1);
ATP binding / ATPase/ nucleoside-triphosphatase/ nucleotide
binding / protein binding
Length=945
Score = 117 bits (293), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 62/137 (45%), Positives = 94/137 (68%), Gaps = 2/137 (1%)
Query 131 VSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQILSRKMTKVPMLVGEPGVGKTAVIEGLA 190
+ Q+ VDL+ A +GL+ PV+GR++E+ R+ QIL R+ P+L+GE GVGKTA+ EGLA
Sbjct 271 LEQFCVDLTARASEGLIDPVIGREKEVQRVIQILCRRTKNNPILLGEAGVGKTAIAEGLA 330
Query 191 QRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQNNVEEVILFVDEI 250
I E + P L +RI +LD+ L AG+ RGE E R+ ++S ++ + +VILF+DE+
Sbjct 331 ISIAEASAPGFLLTKRIMSLDIGLLMAGAKERGELEARVTALISEVKKS-GKVILFIDEV 389
Query 251 HTLIGAAGGVCRNDRAN 267
HTLIG +G V R ++ +
Sbjct 390 HTLIG-SGTVGRGNKGS 405
> tpv:TP04_0800 ATP-dependent Clp protease ATP-binding subunit
(EC:3.4.21.92); K01358 ATP-dependent Clp protease, protease
subunit [EC:3.4.21.92]
Length=900
Score = 114 bits (284), Expect = 5e-25, Method: Composition-based stats.
Identities = 59/137 (43%), Positives = 85/137 (62%), Gaps = 1/137 (0%)
Query 131 VSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQILSRKMTKVPMLVGEPGVGKTAVIEGLA 190
+S + VDL+ A+ G LP V+ RD EI+R LSR P+LVGEPGVGKTA++EG+A
Sbjct 250 ISMFTVDLTEKARNGQLPKVIHRDNEIERAIITLSRMTKSNPLLVGEPGVGKTAIVEGIA 309
Query 191 QRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQNNVEEVILFVDEI 250
RI +G + K+RI L L AG+ RG+FE+R+ +++ ++ + ++IL +DE
Sbjct 310 NRISQGISQPQISKKRILQLQFGLLIAGTKFRGQFEERLTKLIDEIK-SAGDIILVIDEA 368
Query 251 HTLIGAAGGVCRNDRAN 267
H LIG G D AN
Sbjct 369 HMLIGGGAGDGSIDAAN 385
> bbo:BBOV_III008980 17.m07783; Clp amino terminal domain containing
protein; K01358 ATP-dependent Clp protease, protease
subunit [EC:3.4.21.92]
Length=1005
Score = 111 bits (278), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 57/137 (41%), Positives = 90/137 (65%), Gaps = 1/137 (0%)
Query 131 VSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQILSRKMTKVPMLVGEPGVGKTAVIEGLA 190
++ + +D++ A++G L V+ RD EIDR + L RK + P+L+GEPGVGKTAV+EG+A
Sbjct 265 LNAFTIDITRKAEEGKLQKVLCRDSEIDRSIRTLCRKYKRNPILIGEPGVGKTAVVEGIA 324
Query 191 QRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQNNVEEVILFVDEI 250
++ EG V E + +R+ LD+ L AG+ RG+FE+R+ ++ ++ N + +IL +DE
Sbjct 325 MQLREGHVLEKMLNKRLRQLDVGLLVAGARFRGQFEERLTRLIEEIK-NAKNIILVIDEA 383
Query 251 HTLIGAAGGVCRNDRAN 267
H L+GA G D AN
Sbjct 384 HMLVGAGAGEGALDAAN 400
> pfa:PF14_0063 ATP-dependent CLP protease, putative; K03695 ATP-dependent
Clp protease ATP-binding subunit ClpB
Length=1341
Score = 107 bits (268), Expect = 3e-23, Method: Composition-based stats.
Identities = 59/138 (42%), Positives = 87/138 (63%), Gaps = 1/138 (0%)
Query 130 FVSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQILSRKMTKVPMLVGEPGVGKTAVIEGL 189
F+ +D+ AQ+ GR +EI RI +IL RK P+L+GE GVGKTA+IE L
Sbjct 505 FMKDCLIDMVHEAQEKGDDHFFGRKKEIKRIIEILGRKKKSNPLLIGESGVGKTAIIEYL 564
Query 190 AQRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQNNVEEVILFVDE 249
+ I++ VP L+ RIF L+L ++ AG+ RGEFE++MK ++S + N ++ ILF+DE
Sbjct 565 SYLILKDNVPYHLKNCRIFQLNLGNIVAGTKYRGEFEEKMKHLLSNM-NKKKKNILFIDE 623
Query 250 IHTLIGAAGGVCRNDRAN 267
IH ++GA G D +N
Sbjct 624 IHVIVGAGSGEGSLDASN 641
> ath:AT3G45450 Clp amino terminal domain-containing protein
Length=341
Score = 80.1 bits (196), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 45/116 (38%), Positives = 69/116 (59%), Gaps = 9/116 (7%)
Query 142 AQQGLLPPVVGRDEEIDRIAQILSRKMTKV-PMLVGEPGVGKTAVIEGLAQRIVEGAVPE 200
++G L PVVGR +I R+ QIL+R+ + L+G+PGVGK A+ EG+AQRI G VPE
Sbjct 148 TRRGKLDPVVGRQPQIKRVVQILARRTCRNNACLIGKPGVGKRAIAEGIAQRIASGDVPE 207
Query 201 SLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQNNVEEVILFVDEIHTLIGA 256
+++ + AG+ E R + + + +++ILF+DE+H LIGA
Sbjct 208 TIKGKMNV--------AGNCGWNEIRWRSRGKIEEVYGQSDDIILFIDEMHLLIGA 255
> bbo:BBOV_I001700 19.m02115; chaperone clpB
Length=833
Score = 72.4 bits (176), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 44/119 (36%), Positives = 69/119 (57%), Gaps = 9/119 (7%)
Query 137 DLSFMAQQGLLPPVVGRDEEIDRIAQILSRKMTKVPMLVGEPGVGKTAVIEGLA-QRIVE 195
+L+ A+ G VGR+ E++R+ L+R +L+GEPGVGKTA++E LA ++E
Sbjct 168 NLTEAAKNGSGNVFVGRENELERLKGSLNRMRKNNVLLIGEPGVGKTALVERLAVDMLLE 227
Query 196 GAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQNNVEEVILFVDEIHTLI 254
+++LDL L +G RGE E ++K + ++N + ILF+DEIH LI
Sbjct 228 DP------NITVYSLDLCRLYSGQGTRGELEAKLKSIFDTVKNG--KSILFIDEIHHLI 278
> sce:YNL218W MGS1; Mgs1p; K07478 putative ATPase
Length=587
Score = 39.7 bits (91), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 48/169 (28%), Positives = 75/169 (44%), Gaps = 23/169 (13%)
Query 99 EIVEQFKGRQRGVAPTSFQEMVGDKQGTSDAFVSQYGVDLSFMAQQGLLPPVVG-RDEEI 157
EIVE R + T F + + + + D + Y + ++ + LP R +E+
Sbjct 87 EIVENESKRFKAAPSTDFAKSIVEPASSRDQLHNDY--ESRWLQKISHLPLSEKLRPKEL 144
Query 158 -DRIAQ--ILSR---------KMTKVP--MLVGEPGVGKTAVIEGLAQRIVEGAVPESLR 203
D + Q ILS+ K +P +L G PGVGKT++ L + + ES
Sbjct 145 RDYVGQQHILSQDNGTLFKYIKQGTIPSMILWGPPGVGKTSLAR-LLTKTATTSSNESNV 203
Query 204 KRRIFALDLLSLSAGSA-MRGEFEKRMKEVVSYLQNNVEEVILFVDEIH 251
R F ++ + A + +RG FEK KE Q +LF+DEIH
Sbjct 204 GSRYFMIETSATKANTQELRGIFEKSKKEY----QLTKRRTVLFIDEIH 248
> ath:AT1G50140 ATP binding / ATPase/ nucleoside-triphosphatase/
nucleotide binding
Length=981
Score = 38.1 bits (87), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 30/95 (31%), Positives = 46/95 (48%), Gaps = 19/95 (20%)
Query 173 MLVGEPGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSAG---SAMRGEFEKRM 229
+L G PG GKT + + LA GA + +S++ S G+ EK
Sbjct 731 LLFGPPGTGKTLLAKALATEA--GA-------------NFISITGSTLTSKWFGDAEKLT 775
Query 230 KEVVSYLQNNVEEVILFVDEIHTLIGAAGGVCRND 264
K + S+ + VI+FVDEI +L+GA GG ++
Sbjct 776 KALFSF-ATKLAPVIIFVDEIDSLLGARGGSSEHE 809
> ath:AT3G19740 ATP binding / ATPase/ nucleoside-triphosphatase/
nucleotide binding
Length=1001
Score = 37.4 bits (85), Expect = 0.062, Method: Compositional matrix adjust.
Identities = 29/91 (31%), Positives = 45/91 (49%), Gaps = 19/91 (20%)
Query 173 MLVGEPGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSAG---SAMRGEFEKRM 229
+L G PG GKT + + LA GA + +S++ S G+ EK
Sbjct 751 LLFGPPGTGKTLLAKALATEA--GA-------------NFISITGSTLTSKWFGDAEKLT 795
Query 230 KEVVSYLQNNVEEVILFVDEIHTLIGAAGGV 260
K + S+ + + VI+FVDE+ +L+GA GG
Sbjct 796 KALFSF-ASKLAPVIIFVDEVDSLLGARGGA 825
> mmu:69716 Trip13, 2410002G23Rik, D13Ertd328e; thyroid hormone
receptor interactor 13
Length=432
Score = 37.0 bits (84), Expect = 0.074, Method: Compositional matrix adjust.
Identities = 36/143 (25%), Positives = 71/143 (49%), Gaps = 14/143 (9%)
Query 124 QGTSDAFVSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQILSRKMTKVPMLVGEPGVGKT 183
G D+ V Y V++ ++ V+ D+ +D + +++ +V +L G PG GKT
Sbjct 133 HGLWDSLV--YDVEVKSHLLDYVMTTVLFSDKNVD--SNLIT--WNRVVLLHGPPGTGKT 186
Query 184 AVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQNNVEE- 242
++ + LAQ++ + S R R +++ S S S E K + ++ +Q+ +++
Sbjct 187 SLCKALAQKL---TIRLSSRYRYGQLIEINSHSLFSKWFSESGKLVTKMFQKIQDLIDDK 243
Query 243 ---VILFVDEIHTLIGAAGGVCR 262
V + +DE+ +L AA CR
Sbjct 244 EALVFVLIDEVESL-TAARNACR 265
> hsa:9319 TRIP13, 16E1BP; thyroid hormone receptor interactor
13
Length=289
Score = 35.8 bits (81), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 28/99 (28%), Positives = 51/99 (51%), Gaps = 8/99 (8%)
Query 168 MTKVPMLVGEPGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEK 227
+V +L G PG GKT++ + LAQ++ + S R R +++ S S S E K
Sbjct 171 WNRVVLLHGPPGTGKTSLCKALAQKL---TIRLSSRYRYGQLIEINSHSLFSKWFSESGK 227
Query 228 RMKEVVSYLQNNVEE----VILFVDEIHTLIGAAGGVCR 262
+ ++ +Q+ +++ V + +DE+ +L AA CR
Sbjct 228 LVTKMFQKIQDLIDDKDALVFVLIDEVESL-TAARNACR 265
> dre:317678 ruvbl2, reptin, wu:fi25f01, zreptin; RuvB-like 2
(E. coli) (EC:3.6.4.12); K11338 RuvB-like protein 2 [EC:3.6.4.12]
Length=463
Score = 35.0 bits (79), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 37/72 (51%), Gaps = 1/72 (1%)
Query 173 MLVGEPGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSAGS-AMRGEFEKRMKE 231
++ G+PG GKTA+ G+AQ + +L IF+L++ A S A R R+KE
Sbjct 74 LIAGQPGTGKTAIAMGIAQSLGPDTPFTALAGSEIFSLEMSKTEALSQAFRKAIGVRIKE 133
Query 232 VVSYLQNNVEEV 243
++ V E+
Sbjct 134 ETEIIEGEVVEI 145
> hsa:284434 NWD1, MGC134940; NACHT and WD repeat domain containing
1
Length=1432
Score = 34.7 bits (78), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 36/128 (28%), Positives = 58/128 (45%), Gaps = 11/128 (8%)
Query 96 FKHEIVEQFKGRQRGVAPTSFQEMVGDKQGTSDAFVSQYGVDLSFMAQQGLLPPVVGRDE 155
+ E+ EQF R T +E+ D G A++ Q + ++ GR E
Sbjct 261 YLKELGEQFVVRANHQVLTRLREL--DTAGQELAWLYQ-EIRHHLWQSSEVIQTFCGRQE 317
Query 156 EIDRIAQILSRKMTK--VPM-LVGEPGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDL 212
+ R+ Q L +K P+ L G PG+GKTA++ LA+++ P L + + L L
Sbjct 318 LLARLGQQLRHDDSKQHTPLVLFGPPGIGKTALMCKLAEQM-----PRLLGHKTVTVLRL 372
Query 213 LSLSAGSA 220
L S S+
Sbjct 373 LGTSQMSS 380
> tgo:TGME49_069890 M16 family peptidase, putative (EC:3.4.24.56)
Length=941
Score = 34.3 bits (77), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 34/74 (45%), Gaps = 6/74 (8%)
Query 104 FKGRQRGVAPTSFQEMVGDKQGTSDAFVSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQI 163
F+G +R F + V + G ++AF S++ SF G L P +DR+A +
Sbjct 86 FQGSKRFPGTHDFFDFVHNHGGYTNAFTSKFSTVFSFSIGPGFLEP------GLDRLADL 139
Query 164 LSRKMTKVPMLVGE 177
S + K L+ E
Sbjct 140 FSAPLLKSENLLKE 153
> sce:YOL094C RFC4; Rfc4p; K10755 replication factor C subunit
2/4
Length=323
Score = 34.3 bits (77), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 26/49 (53%), Gaps = 0/49 (0%)
Query 146 LLPPVVGRDEEIDRIAQILSRKMTKVPMLVGEPGVGKTAVIEGLAQRIV 194
+L +VG E IDR+ QI ++ G PG+GKT + LA ++
Sbjct 19 VLSDIVGNKETIDRLQQIAKDGNMPHMIISGMPGIGKTTSVHCLAHELL 67
> tpv:TP03_0470 origin recognition complex 1 protein
Length=645
Score = 34.3 bits (77), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 25/131 (19%), Positives = 57/131 (43%), Gaps = 17/131 (12%)
Query 150 VVGRDEEIDRIAQILSRKMTK-----VPMLVGEPGVGKTAVIEGLAQRIVEGAVPESLRK 204
++GR++E D+I + + + V + G PG GKT ++ +++ ++ + +
Sbjct 256 ILGREDEADKIRTFMETNIKQGGTGQVLYISGVPGTGKTETVKMVSKELIGKKLKGQIPW 315
Query 205 RRIFALDLLSLSAGSAMRGEFEKRM------------KEVVSYLQNNVEEVILFVDEIHT 252
+ ++ + LS + + F ++ +E+ Y NN+ +L VDE
Sbjct 316 FDLVEINAVHLSRPNELYRVFYNKLFAKPAPASHICYEELDKYFTNNMTPCVLIVDEADY 375
Query 253 LIGAAGGVCRN 263
++ V N
Sbjct 376 IVTKTQKVLFN 386
> ath:AT4G02480 AAA-type ATPase family protein
Length=1265
Score = 33.9 bits (76), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 28/89 (31%), Positives = 45/89 (50%), Gaps = 13/89 (14%)
Query 167 KMTKVPMLVGEPGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFE 226
K TK +L G PG GKT + + +A GA + + S++ S GE E
Sbjct 996 KPTKGILLFGPPGTGKTMLAKAVATEA--GA--------NFINISMSSIT--SKWFGEGE 1043
Query 227 KRMKEVVSYLQNNVEEVILFVDEIHTLIG 255
K +K V S L + + ++FVDE+ +++G
Sbjct 1044 KYVKAVFS-LASKIAPSVIFVDEVDSMLG 1071
> xla:100037172 trip13; thyroid hormone receptor interactor 13
Length=352
Score = 33.9 bits (76), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 28/93 (30%), Positives = 49/93 (52%), Gaps = 7/93 (7%)
Query 168 MTKVPMLVGEPGVGKTAVIEGLAQRIVEGAVPESLRKR--RIFALDLLSL-SAGSAMRGE 224
+V +L G PG GKT++ + LAQ++ V S R R ++ ++ SL S + G+
Sbjct 171 WNRVVLLHGPPGTGKTSLCKALAQKL---TVRLSYRYRYGQLVEINSHSLFSKWFSESGK 227
Query 225 FEKRMKEVVSYLQNNVEE-VILFVDEIHTLIGA 256
+M + + L N+ E V + +DE+ +L A
Sbjct 228 LVTKMFQKIHDLINDKEALVFVLIDEVESLTAA 260
> xla:380092 ruvbl2, MGC52995; RuvB-like protein 2; K11338 RuvB-like
protein 2 [EC:3.6.4.12]
Length=462
Score = 33.9 bits (76), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 36/72 (50%), Gaps = 1/72 (1%)
Query 173 MLVGEPGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSA-GSAMRGEFEKRMKE 231
++ G+PG GKTA+ G+AQ + ++ IF+L++ A A R R+KE
Sbjct 73 LIAGQPGTGKTAIAMGMAQALGSDTPFTAIAGSEIFSLEMSKTEALTQAFRRSIGVRIKE 132
Query 232 VVSYLQNNVEEV 243
++ V EV
Sbjct 133 ETEIIEGEVVEV 144
> xla:398205 ruvbl2, tip48, xReptin; RuvB-like 2 (EC:3.6.4.12);
K11338 RuvB-like protein 2 [EC:3.6.4.12]
Length=462
Score = 33.9 bits (76), Expect = 0.60, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 36/72 (50%), Gaps = 1/72 (1%)
Query 173 MLVGEPGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSA-GSAMRGEFEKRMKE 231
++ G+PG GKTA+ G+AQ + ++ IF+L++ A A R R+KE
Sbjct 73 LIAGQPGTGKTAIAMGMAQALGSDTPFTAIAGSEIFSLEMSKTEALTQAFRRSIGVRIKE 132
Query 232 VVSYLQNNVEEV 243
++ V EV
Sbjct 133 ETEIIEGEVVEV 144
> dre:405856 MGC85976; zgc:85976; K07478 putative ATPase
Length=546
Score = 33.9 bits (76), Expect = 0.68, Method: Compositional matrix adjust.
Identities = 27/95 (28%), Positives = 44/95 (46%), Gaps = 16/95 (16%)
Query 162 QILSRKMTK---VPMLV--GEPGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLS 216
Q L R + K +P L+ G PG GKT + +A I + ++LS
Sbjct 139 QTLLRSLLKSQEIPSLILWGPPGCGKTTLAHIIASSIKQKGTGR-----------FVTLS 187
Query 217 AGSAMRGEFEKRMKEVVSYLQNNVEEVILFVDEIH 251
A SA + + +K+ + L+ + +LF+DEIH
Sbjct 188 ATSASVSDVREVIKQAQNELRLCKRKTVLFIDEIH 222
> xla:734231 wrnip1; Werner helicase interacting protein 1; K07478
putative ATPase
Length=572
Score = 33.5 bits (75), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 38/79 (48%), Gaps = 12/79 (15%)
Query 173 MLVGEPGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEV 232
+L G PG GKT LA I + A S R ++LSA SA + + +K+
Sbjct 188 ILWGPPGCGKTT----LAHIIAKNAQKSSSR--------FVTLSATSASTSDVREFIKQA 235
Query 233 VSYLQNNVEEVILFVDEIH 251
+ + + ILFVDEIH
Sbjct 236 QNEQRLFKRKTILFVDEIH 254
> ath:AT4G24710 ATP binding / ATPase/ nucleoside-triphosphatase/
nucleotide binding
Length=475
Score = 33.5 bits (75), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 47/97 (48%), Gaps = 15/97 (15%)
Query 168 MTKVPMLVGEPGVGKTAVIEGLAQRIVEGAVPESLR-KRRIFALDLLSLSAGSAMRGEFE 226
++ +L G PG GKT++ + LAQ++ S+R R L+ ++A S F
Sbjct 209 WNRIILLHGPPGTGKTSLCKALAQKL-------SIRCNSRYPHCQLIEVNAHSLFSKWFS 261
Query 227 ---KRMKEVVSYLQNNVEE----VILFVDEIHTLIGA 256
K + ++ +Q VEE V + +DE+ +L A
Sbjct 262 ESGKLVAKLFQKIQEMVEEDGNLVFVLIDEVESLAAA 298
> dre:563001 hypothetical LOC563001
Length=3148
Score = 33.5 bits (75), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 52/256 (20%), Positives = 105/256 (41%), Gaps = 41/256 (16%)
Query 19 FIRHSVRDFIVGLPRSSVPINLGESGPSVALRN-------ALRKAWKMAEFEGAPIGVQH 71
F+ H RDF +S I++ + PS +N + RK W+ H
Sbjct 959 FMIHMARDF------ASPSIDISDQSPSFFSKNEDEEEILSFRKRWENESHPYIFFNADH 1012
Query 72 LYLALVEILAFQKIMVKSGCDLR----LFKHEIVEQ-FKGRQRGVAPTS--FQEMVGDKQ 124
+ ++ + Q + + D + L ++ + ++ F QR + S F ++ + +
Sbjct 1013 VSMSFLGFHVKQNGTILNAVDSKSGKVLMRNVMTQELFSDIQRQMINLSKDFDDLTREDK 1072
Query 125 GTSDAFVSQYGVDLSFMAQQGLLPPVVGRDEEIDRIAQILSRKM---TKVPMLV-GEPGV 180
+FV V ++G P + D + ++L+ M ++P+++ GE G
Sbjct 1073 LQKMSFV----VGAEKGCEKGKFDPDPTYELTTDNVMKMLAIHMRFRCEIPVIIMGETGC 1128
Query 181 GKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQNNV 240
GKT ++ L EG R + + L+ + G+ + K+++E Q N
Sbjct 1129 GKTRLVRFLCDLQREG--------RDVENMKLVKVHGGTTSETIY-KKVREAEELAQKNR 1179
Query 241 E----EVILFVDEIHT 252
+ + +LF DE +T
Sbjct 1180 QKYKLDTVLFFDEANT 1195
> bbo:BBOV_III003410 17.m10506; hypothetical protein
Length=195
Score = 33.5 bits (75), Expect = 0.90, Method: Compositional matrix adjust.
Identities = 25/113 (22%), Positives = 51/113 (45%), Gaps = 14/113 (12%)
Query 147 LPPVVGRDEEIDRIAQILSRKMTKVPMLVGEPGVGKTAVIEGLAQR----IVEGAVPESL 202
+P + ++ E+D Q ++ + K+ + + E V K EG R IV+G++PE L
Sbjct 44 VPSLKSKEGEVDNDMQWVTHRKVKMVLWLSETSVQKEIANEGEVARALVYIVDGSIPEKL 103
Query 203 RKRRIFALDLLSLSAGSAMRGEFEKRMKEVVSYLQNNVEEVILFVDEIHTLIG 255
+ + + L +AG+ ++ L + +F++E H +G
Sbjct 104 AEAKEYLKQQLEQTAGAP----------NLLILLNKCDKHSFIFLEEAHDALG 146
> cpv:cgd6_550 Pch2p like AAA ATpase
Length=546
Score = 33.1 bits (74), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 24/88 (27%), Positives = 47/88 (53%), Gaps = 7/88 (7%)
Query 173 MLVGEPGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEV 232
+L G PG GKT++ ++Q+I + R + I+ L++ + S S E K + ++
Sbjct 278 LLYGSPGTGKTSISRAISQKI---GMRYCHRYKNIYLLEISAHSLFSKWFSESGKTVVKL 334
Query 233 VSYLQNNVEE----VILFVDEIHTLIGA 256
S +++ +EE V + +DEI ++ A
Sbjct 335 FSKIKSLLEEPDSFVNIVIDEIESISTA 362
> ath:AT4G24860 AAA-type ATPase family protein
Length=1122
Score = 33.1 bits (74), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 24/83 (28%), Positives = 40/83 (48%), Gaps = 13/83 (15%)
Query 173 MLVGEPGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSAGSAMRGEFEKRMKEV 232
+L G PG GKT + + +A+ + S+ S S GE EK +K V
Sbjct 859 LLFGPPGTGKTMLAKAVAKEADANFINISMS------------SITSKWFGEGEKYVKAV 906
Query 233 VSYLQNNVEEVILFVDEIHTLIG 255
S L + + ++FVDE+ +++G
Sbjct 907 FS-LASKMSPSVIFVDEVDSMLG 928
> dre:100170780 zgc:195077
Length=522
Score = 33.1 bits (74), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 165 SRKMTKVPMLVGEPGVGKTAVIEGLAQRIV 194
+RK K+ +LVGE G GKT +I L I+
Sbjct 41 ARKQNKIILLVGETGTGKTTLINSLVNYIL 70
> mmu:319555 Nwd1, A230063L24Rik; NACHT and WD repeat domain containing
1
Length=1521
Score = 33.1 bits (74), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 32/56 (57%), Gaps = 1/56 (1%)
Query 169 TKVPM-LVGEPGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSAGSAMRG 223
T P+ L G PG+GKT+++ LAQ++ E +++ R+ LSL A S +R
Sbjct 334 THTPLVLFGPPGIGKTSLMCKLAQQVPELLGHKTVVVLRLLGTSKLSLDARSLLRS 389
> mmu:20174 Ruvbl2, MGC144733, MGC144734, mp47, p47; RuvB-like
protein 2 (EC:3.6.4.12); K11338 RuvB-like protein 2 [EC:3.6.4.12]
Length=463
Score = 32.7 bits (73), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 36/72 (50%), Gaps = 1/72 (1%)
Query 173 MLVGEPGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSA-GSAMRGEFEKRMKE 231
++ G+PG GKTA+ G+AQ + ++ IF+L++ A A R R+KE
Sbjct 74 LIAGQPGTGKTAIAMGMAQALGPDTPFTAIAGSEIFSLEMSKTEALTQAFRRSIGVRIKE 133
Query 232 VVSYLQNNVEEV 243
++ V E+
Sbjct 134 ETEIIEGEVVEI 145
> hsa:10856 RUVBL2, ECP51, INO80J, REPTIN, RVB2, TIH2, TIP48,
TIP49B; RuvB-like 2 (E. coli) (EC:3.6.4.12); K11338 RuvB-like
protein 2 [EC:3.6.4.12]
Length=463
Score = 32.7 bits (73), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 36/72 (50%), Gaps = 1/72 (1%)
Query 173 MLVGEPGVGKTAVIEGLAQRIVEGAVPESLRKRRIFALDLLSLSA-GSAMRGEFEKRMKE 231
++ G+PG GKTA+ G+AQ + ++ IF+L++ A A R R+KE
Sbjct 74 LIAGQPGTGKTAIAMGMAQALGPDTPFTAIAGSEIFSLEMSKTEALTQAFRRSIGVRIKE 133
Query 232 VVSYLQNNVEEV 243
++ V E+
Sbjct 134 ETEIIEGEVVEI 145
Lambda K H
0.321 0.138 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9954723792
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40