bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1830_orf1
Length=211
Score E
Sequences producing significant alignments: (Bits) Value
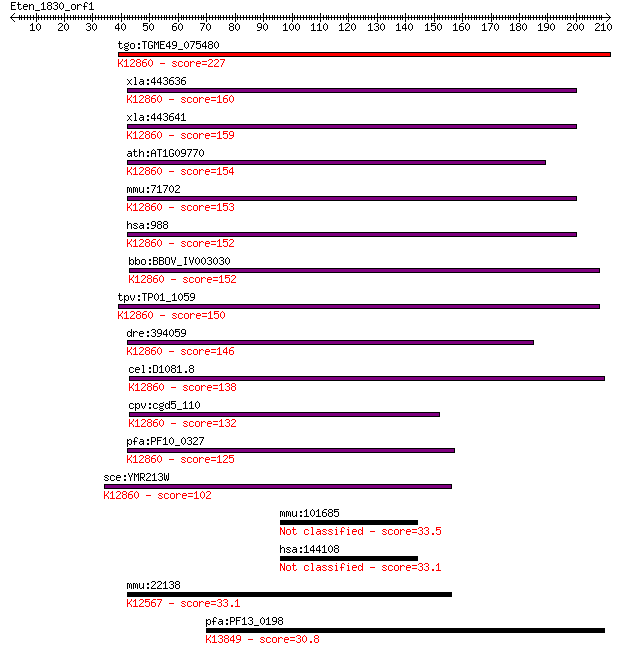
tgo:TGME49_075480 myb-like DNA-binding domain-containing prote... 227 2e-59
xla:443636 cdc5l, MGC154633; CDC5 cell division cycle 5-like; ... 160 4e-39
xla:443641 cdc5l, MGC114655; cell division cycle 5-like; K1286... 159 6e-39
ath:AT1G09770 ATCDC5; ATCDC5 (ARABIDOPSIS THALIANA CELL DIVISI... 154 2e-37
mmu:71702 Cdc5l, 1200002I02Rik, AA408004, PCDC5RP; cell divisi... 153 4e-37
hsa:988 CDC5L, CDC5, CDC5-LIKE, CEF1, KIAA0432, PCDC5RP, dJ319... 152 6e-37
bbo:BBOV_IV003030 21.m02918; cell division cycle 5-like protei... 152 8e-37
tpv:TP01_1059 hypothetical protein; K12860 pre-mRNA-splicing f... 150 3e-36
dre:394059 cdc5l, MGC55853, zgc:55853; CDC5 cell division cycl... 146 4e-35
cel:D1081.8 hypothetical protein; K12860 pre-mRNA-splicing fac... 138 1e-32
cpv:cgd5_110 CDC5 cell division cycle 5-like ; K12860 pre-mRNA... 132 6e-31
pfa:PF10_0327 Myb2 protein; K12860 pre-mRNA-splicing factor CD... 125 1e-28
sce:YMR213W CEF1, NTC85; Cef1p; K12860 pre-mRNA-splicing facto... 102 8e-22
mmu:101685 Spty2d1, 5830435K17Rik, AI852426, P16H6; SPT2, Supp... 33.5 0.62
hsa:144108 SPTY2D1, DKFZp686F1942, DKFZp686I068, FLJ39441; SPT... 33.1 0.63
mmu:22138 Ttn, 1100001C23Rik, 2310036G12Rik, 2310057K23Rik, 23... 33.1 0.67
pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog A... 30.8 3.5
> tgo:TGME49_075480 myb-like DNA-binding domain-containing protein
; K12860 pre-mRNA-splicing factor CDC5/CEF1
Length=888
Score = 227 bits (579), Expect = 2e-59, Method: Compositional matrix adjust.
Identities = 130/175 (74%), Positives = 151/175 (86%), Gaps = 2/175 (1%)
Query 39 RGR-EDPRRLRPGEIDPHPETKPSRADPIDMAEDEKEMLEEARARLANTRGKKAKRKARE 97
RGR +DPRRLRPGEIDPHPETKPSRAD IDM +DEKEML+EARARLANTRGKKAKRKARE
Sbjct 137 RGRFDDPRRLRPGEIDPHPETKPSRADAIDMQDDEKEMLQEARARLANTRGKKAKRKARE 196
Query 98 KQLEEARRLAALQKKRELKAAGIITGAKRRTRKNFQPY-EEVPFEEKPPPGFYEVPSEEN 156
KQLEEARRLA+LQK+RE+KAAG+IT + R+ Y ++PFEEKPPPGF+ V +EE
Sbjct 197 KQLEEARRLASLQKRREMKAAGLITSLRPHKRRREMDYGVDIPFEEKPPPGFHAVGAEET 256
Query 157 PEGNLNFANISLQHMEGSMRAREEEKLRKEDARKLKRLREDHLDEYLKIHEEKLK 211
PEGNLNFANISLQ +EG+MRA+EE KLR+EDARKLKRL+E++L YL+ EEK K
Sbjct 257 PEGNLNFANISLQQLEGTMRAQEEMKLRREDARKLKRLQEENLPAYLQQLEEKNK 311
> xla:443636 cdc5l, MGC154633; CDC5 cell division cycle 5-like;
K12860 pre-mRNA-splicing factor CDC5/CEF1
Length=804
Score = 160 bits (404), Expect = 4e-39, Method: Compositional matrix adjust.
Identities = 96/159 (60%), Positives = 123/159 (77%), Gaps = 2/159 (1%)
Query 42 EDPRRLRPGEIDPHPETKPSRADPIDMAEDEKEMLEEARARLANTRGKKAKRKAREKQLE 101
+DPR+L+PGEIDP+PETKP+R DP+DM EDE EML EARARLANT+GKKAKRKAREKQLE
Sbjct 118 DDPRKLKPGEIDPNPETKPARPDPVDMDEDELEMLSEARARLANTQGKKAKRKAREKQLE 177
Query 102 EARRLAALQKKRELKAAGIITGAKRRTRKNFQPYEEVPFEEKPPPGFYEVPSEENPEG-N 160
EARRLAALQK+REL+AAGI KR+ ++ E+PFE+KP PGFY+ SEEN + N
Sbjct 178 EARRLAALQKRRELRAAGIDIQKKRKKKRGVDYNAEIPFEKKPAPGFYDT-SEENYDALN 236
Query 161 LNFANISLQHMEGSMRAREEEKLRKEDARKLKRLREDHL 199
+F + Q ++G +R+ +E K RK+D + +KR +E L
Sbjct 237 ADFRKLRQQDLDGDLRSEKEAKDRKKDKQNIKRKKESDL 275
> xla:443641 cdc5l, MGC114655; cell division cycle 5-like; K12860
pre-mRNA-splicing factor CDC5/CEF1
Length=804
Score = 159 bits (402), Expect = 6e-39, Method: Compositional matrix adjust.
Identities = 93/158 (58%), Positives = 119/158 (75%), Gaps = 0/158 (0%)
Query 42 EDPRRLRPGEIDPHPETKPSRADPIDMAEDEKEMLEEARARLANTRGKKAKRKAREKQLE 101
+DPR+L+PGEIDP+PETKP+R DP+DM EDE EML EARARLANT+GKKAKRKAREKQLE
Sbjct 118 DDPRKLKPGEIDPNPETKPARPDPVDMDEDELEMLSEARARLANTQGKKAKRKAREKQLE 177
Query 102 EARRLAALQKKRELKAAGIITGAKRRTRKNFQPYEEVPFEEKPPPGFYEVPSEENPEGNL 161
EARRLAALQK+REL+AAGI KR+ ++ E+PFE+KP PGFY+ E N
Sbjct 178 EARRLAALQKRRELRAAGIEIQKKRKKKRGVDYNAEIPFEKKPAPGFYDTSEENYNALNA 237
Query 162 NFANISLQHMEGSMRAREEEKLRKEDARKLKRLREDHL 199
+F + Q ++G +R+ +E K RK+D + +KR +E L
Sbjct 238 DFRKLRQQDLDGDLRSEKEAKDRKKDKQNIKRKKESDL 275
> ath:AT1G09770 ATCDC5; ATCDC5 (ARABIDOPSIS THALIANA CELL DIVISION
CYCLE 5); DNA binding / transcription factor; K12860 pre-mRNA-splicing
factor CDC5/CEF1
Length=844
Score = 154 bits (389), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 91/147 (61%), Positives = 111/147 (75%), Gaps = 1/147 (0%)
Query 42 EDPRRLRPGEIDPHPETKPSRADPIDMAEDEKEMLEEARARLANTRGKKAKRKAREKQLE 101
+DPR+LRPGEIDP+PE KP+R DP+DM EDEKEML EARARLANTRGKKAKRKAREKQLE
Sbjct 117 DDPRKLRPGEIDPNPEAKPARPDPVDMDEDEKEMLSEARARLANTRGKKAKRKAREKQLE 176
Query 102 EARRLAALQKKRELKAAGIITGAKRRTRKNFQPYEEVPFEEKPPPGFYEVPSEENPEGNL 161
EARRLA+LQK+RELKAAGI ++R RK E+PFE++ P GFY+ E+ P +
Sbjct 177 EARRLASLQKRRELKAAGIDGRHRKRKRKGIDYNAEIPFEKRAPAGFYDTADEDRPADQV 236
Query 162 NFANISLQHMEGSMRAREEEKLRKEDA 188
F +++ +EG RA E LRK+D
Sbjct 237 KFPT-TIEELEGKRRADVEAHLRKQDV 262
> mmu:71702 Cdc5l, 1200002I02Rik, AA408004, PCDC5RP; cell division
cycle 5-like (S. pombe); K12860 pre-mRNA-splicing factor
CDC5/CEF1
Length=802
Score = 153 bits (386), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 95/159 (59%), Positives = 122/159 (76%), Gaps = 2/159 (1%)
Query 42 EDPRRLRPGEIDPHPETKPSRADPIDMAEDEKEMLEEARARLANTRGKKAKRKAREKQLE 101
+DPR+L+PGEIDP+PETKP+R DPIDM EDE EML EARARLANT+GKKAKRKAREKQLE
Sbjct 118 DDPRKLKPGEIDPNPETKPARPDPIDMDEDELEMLSEARARLANTQGKKAKRKAREKQLE 177
Query 102 EARRLAALQKKRELKAAGIITGAKRRTRKNFQPYEEVPFEEKPPPGFYEVPSEENPEG-N 160
EARRLAALQK+REL+AAGI KR+ ++ E+PFE+KP GFY+ SEEN + +
Sbjct 178 EARRLAALQKRRELRAAGIEIQKKRKKKRGVDYNAEIPFEKKPALGFYDT-SEENYQALD 236
Query 161 LNFANISLQHMEGSMRAREEEKLRKEDARKLKRLREDHL 199
+F + Q ++G +R+ +E + RK+D + LKR +E L
Sbjct 237 ADFRKLRQQDLDGELRSEKEGRDRKKDKQHLKRKKESDL 275
> hsa:988 CDC5L, CDC5, CDC5-LIKE, CEF1, KIAA0432, PCDC5RP, dJ319D22.1;
CDC5 cell division cycle 5-like (S. pombe); K12860
pre-mRNA-splicing factor CDC5/CEF1
Length=802
Score = 152 bits (385), Expect = 6e-37, Method: Compositional matrix adjust.
Identities = 95/159 (59%), Positives = 122/159 (76%), Gaps = 2/159 (1%)
Query 42 EDPRRLRPGEIDPHPETKPSRADPIDMAEDEKEMLEEARARLANTRGKKAKRKAREKQLE 101
+DPR+L+PGEIDP+PETKP+R DPIDM EDE EML EARARLANT+GKKAKRKAREKQLE
Sbjct 118 DDPRKLKPGEIDPNPETKPARPDPIDMDEDELEMLSEARARLANTQGKKAKRKAREKQLE 177
Query 102 EARRLAALQKKRELKAAGIITGAKRRTRKNFQPYEEVPFEEKPPPGFYEVPSEENPEG-N 160
EARRLAALQK+REL+AAGI KR+ ++ E+PFE+KP GFY+ SEEN + +
Sbjct 178 EARRLAALQKRRELRAAGIEIQKKRKRKRGVDYNAEIPFEKKPALGFYDT-SEENYQALD 236
Query 161 LNFANISLQHMEGSMRAREEEKLRKEDARKLKRLREDHL 199
+F + Q ++G +R+ +E + RK+D + LKR +E L
Sbjct 237 ADFRKLRQQDLDGELRSEKEGRDRKKDKQHLKRKKESDL 275
> bbo:BBOV_IV003030 21.m02918; cell division cycle 5-like protein;
K12860 pre-mRNA-splicing factor CDC5/CEF1
Length=596
Score = 152 bits (384), Expect = 8e-37, Method: Compositional matrix adjust.
Identities = 95/165 (57%), Positives = 121/165 (73%), Gaps = 4/165 (2%)
Query 43 DPRRLRPGEIDPHPETKPSRADPIDMAEDEKEMLEEARARLANTRGKKAKRKAREKQLEE 102
DPRRLRPGEIDP PETKPSRAD +DM +DEKEML EARARLANTRGKKAKRKAREKQ+E+
Sbjct 116 DPRRLRPGEIDPAPETKPSRADAVDMDDDEKEMLAEARARLANTRGKKAKRKAREKQIEQ 175
Query 103 ARRLAALQKKRELKAAGIITGAKRRTRKNFQPYEEVPFEEKPPPGFYEVPSEENPEGNLN 162
RRLA+LQK+ ELK AG+ GA + + +EVPFE +PP GFYE + + +L
Sbjct 176 TRRLASLQKRHELKKAGVNVGALKLHKSIMDYVKEVPFETQPPKGFYEPETGHEIDQSLR 235
Query 163 FANISLQHMEGSMRAREEEKLRKEDARKLKRLREDHLDEYLKIHE 207
S+Q +EG R E ++R +DARKLKRL+E+++ E + + +
Sbjct 236 ----SIQQLEGKRRDDEMRRMRNDDARKLKRLQEENIGEAMAVFQ 276
> tpv:TP01_1059 hypothetical protein; K12860 pre-mRNA-splicing
factor CDC5/CEF1
Length=658
Score = 150 bits (379), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 99/174 (56%), Positives = 121/174 (69%), Gaps = 9/174 (5%)
Query 39 RGRE-----DPRRLRPGEIDPHPETKPSRADPIDMAEDEKEMLEEARARLANTRGKKAKR 93
+GRE DPR+LRPGEIDP E KPSRAD +DM +DEKEML EARARLANTRGKKAKR
Sbjct 107 QGREEDDQFDPRKLRPGEIDPQLECKPSRADAVDMDDDEKEMLAEARARLANTRGKKAKR 166
Query 94 KAREKQLEEARRLAALQKKRELKAAGIITGAKRRTRKNFQPYEEVPFEEKPPPGFYEVPS 153
KAREK LE+++R+A LQK+RELK+AGI + + +E+PFE +PP GFY P
Sbjct 167 KAREKALEQSKRIAMLQKRRELKSAGINVRNFKMKKITMDYEKEIPFEMQPPKGFYP-PD 225
Query 154 EENPEGNLNFANISLQHMEGSMRAREEEKLRKEDARKLKRLREDHLDEYLKIHE 207
EE P NL+ NI + +EG R +E KLRK+D RKLKRL+ D + I E
Sbjct 226 EERP-ANLSIKNI--EQLEGIRRDQEMNKLRKDDIRKLKRLQNDDTPAAMSIFE 276
> dre:394059 cdc5l, MGC55853, zgc:55853; CDC5 cell division cycle
5-like (S. pombe); K12860 pre-mRNA-splicing factor CDC5/CEF1
Length=800
Score = 146 bits (369), Expect = 4e-35, Method: Compositional matrix adjust.
Identities = 85/143 (59%), Positives = 110/143 (76%), Gaps = 0/143 (0%)
Query 42 EDPRRLRPGEIDPHPETKPSRADPIDMAEDEKEMLEEARARLANTRGKKAKRKAREKQLE 101
+DPR+L+PGEIDP+PETKP+R DP+DM EDE EML EARARLANT+GKKAKRKAREKQLE
Sbjct 118 DDPRKLKPGEIDPNPETKPARPDPVDMDEDELEMLSEARARLANTQGKKAKRKAREKQLE 177
Query 102 EARRLAALQKKRELKAAGIITGAKRRTRKNFQPYEEVPFEEKPPPGFYEVPSEENPEGNL 161
EARRLAALQK+REL+AAGI KR+ ++ E+PFE+KP GFY+ E+
Sbjct 178 EARRLAALQKRRELRAAGIDIQKKRKKKRGVDYNAEIPFEKKPAQGFYDTSMEQYDPLEP 237
Query 162 NFANISLQHMEGSMRAREEEKLR 184
+F + QH++G +R+ +E++ R
Sbjct 238 DFKRLRQQHLDGELRSEKEDRDR 260
> cel:D1081.8 hypothetical protein; K12860 pre-mRNA-splicing factor
CDC5/CEF1
Length=755
Score = 138 bits (348), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 89/170 (52%), Positives = 116/170 (68%), Gaps = 6/170 (3%)
Query 43 DPRRLRPGEIDPHPETKPSRADPIDMAEDEKEMLEEARARLANTRGKKAKRKAREKQLEE 102
+ R+L+PGEIDP PETKP+R DPIDM +DE EML EARARLANT+GKKAKRKARE+QL +
Sbjct 120 ETRKLKPGEIDPTPETKPARPDPIDMDDDELEMLSEARARLANTQGKKAKRKARERQLSD 179
Query 103 ARRLAALQKKRELKAAGIITGAKRRTRKNFQPY-EEVPFEEKPPPGFYEVPSEENPEGNL 161
ARRLA+LQK+RE++AAG+ K + ++N Y EE+PFE+ P GF+ PSE+ +
Sbjct 180 ARRLASLQKRREMRAAGLAFARKFKPKRNQIDYSEEIPFEKHVPAGFHN-PSEDR--YVV 236
Query 162 NFANISLQHMEGSMRARE-EEKLRKEDARKLKRLRED-HLDEYLKIHEEK 209
AN R RE E ++R+ED KLK+ +E D I E+K
Sbjct 237 EDANQKAIEDHQKPRGREIEMEMRREDREKLKKRKEQGEADAVFNIKEKK 286
> cpv:cgd5_110 CDC5 cell division cycle 5-like ; K12860 pre-mRNA-splicing
factor CDC5/CEF1
Length=800
Score = 132 bits (333), Expect = 6e-31, Method: Compositional matrix adjust.
Identities = 69/114 (60%), Positives = 87/114 (76%), Gaps = 5/114 (4%)
Query 43 DPRRLRPGEIDPHPETKPSRADPIDMAEDEKEMLEEARARLANTRGKKAKRKAREKQLEE 102
DPR L+PGEIDP+PETKPS+ D IDM E+E EML EARARLANT G+KAKRKARE+ LEE
Sbjct 119 DPRLLKPGEIDPNPETKPSKPDSIDMDEEEIEMLAEARARLANTNGRKAKRKARERYLEE 178
Query 103 ARRLAALQKKRELKAAGIITGA-----KRRTRKNFQPYEEVPFEEKPPPGFYEV 151
ARR+A LQK+RELKAAG+++ A +++ K E+PFEE P G +++
Sbjct 179 ARRIAMLQKRRELKAAGMLSHASIMRYRKKKYKGVDYLNEIPFEEAPEEGAFKM 232
> pfa:PF10_0327 Myb2 protein; K12860 pre-mRNA-splicing factor
CDC5/CEF1
Length=915
Score = 125 bits (313), Expect = 1e-28, Method: Composition-based stats.
Identities = 78/116 (67%), Positives = 93/116 (80%), Gaps = 1/116 (0%)
Query 42 EDPRRLRPGEIDPHPETKPSRADPIDMAEDEKEMLEEARARLANTRGKKAKRKAREKQLE 101
++PR LRPGEIDP PE+KP+RADP+DM EDEKEML EA+ARLANT+GKKAKRKAREKQLE
Sbjct 115 KNPRHLRPGEIDPAPESKPARADPVDMDEDEKEMLAEAKARLANTKGKKAKRKAREKQLE 174
Query 102 EARRLAALQKKRELKAAGIIT-GAKRRTRKNFQPYEEVPFEEKPPPGFYEVPSEEN 156
+ARRLA LQKKRELKAAGI + KR+ + +E+ F KP GFY+V E+N
Sbjct 175 QARRLALLQKKRELKAAGITSLNYKRKDKNKIDHSKEILFHRKPLKGFYDVKDEQN 230
> sce:YMR213W CEF1, NTC85; Cef1p; K12860 pre-mRNA-splicing factor
CDC5/CEF1
Length=590
Score = 102 bits (255), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 57/125 (45%), Positives = 82/125 (65%), Gaps = 6/125 (4%)
Query 34 GVGFLRGREDPRRLRPGEIDPHPETKPSRADPIDMAEDEKEMLEEARARLANTRGKKAKR 93
G G D L+ G+I+P+ ET+ +R D D+ ++EKEML EARARL NT+GKKA R
Sbjct 113 GAALSTGVTD---LKAGDINPNAETQMARPDNGDLEDEEKEMLAEARARLLNTQGKKATR 169
Query 94 KAREKQLEEARRLAALQKKRELKAAGIITGAKRRTRK---NFQPYEEVPFEEKPPPGFYE 150
K RE+ LEE++R+A LQK+RELK AGI K+ +K + E++ +E+ P PG Y+
Sbjct 170 KIRERMLEESKRIAELQKRRELKQAGINVAIKKPKKKYGTDIDYNEDIVYEQAPMPGIYD 229
Query 151 VPSEE 155
+E+
Sbjct 230 TSTED 234
> mmu:101685 Spty2d1, 5830435K17Rik, AI852426, P16H6; SPT2, Suppressor
of Ty, domain containing 1 (S. cerevisiae)
Length=682
Score = 33.5 bits (75), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 22/49 (44%), Positives = 28/49 (57%), Gaps = 1/49 (2%)
Query 96 REKQLEEARRLAALQKKR-ELKAAGIITGAKRRTRKNFQPYEEVPFEEK 143
R+K LEE +R L KKR ELK +RT+ NF Y+ +P EEK
Sbjct 56 RQKALEEKKRKEELVKKRIELKHDKKARAMAKRTKDNFHGYDGIPVEEK 104
> hsa:144108 SPTY2D1, DKFZp686F1942, DKFZp686I068, FLJ39441; SPT2,
Suppressor of Ty, domain containing 1 (S. cerevisiae)
Length=685
Score = 33.1 bits (74), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 23/49 (46%), Positives = 26/49 (53%), Gaps = 1/49 (2%)
Query 96 REKQLEEARRLAALQKKR-ELKAAGIITGAKRRTRKNFQPYEEVPFEEK 143
R K LEE RR L KKR ELK +RT+ NF Y +P EEK
Sbjct 56 RRKALEEKRRKEELVKKRIELKHDKKARAMAKRTKDNFHGYNGIPIEEK 104
> mmu:22138 Ttn, 1100001C23Rik, 2310036G12Rik, 2310057K23Rik,
2310074I15Rik, AF006999, AV006427, D330041I19Rik, D830007G01Rik,
L56, mdm, shru; titin (EC:2.7.11.1); K12567 titin [EC:2.7.11.1]
Length=33467
Score = 33.1 bits (74), Expect = 0.67, Method: Composition-based stats.
Identities = 30/118 (25%), Positives = 52/118 (44%), Gaps = 5/118 (4%)
Query 42 EDPRRLRPGEIDPHPETKPSRADPIDMAEDEKEMLEEARARLANTRGKKAKRKAR----E 97
E+ +R P + P E P +AE ++ +E + +A T+ K+A +AR
Sbjct 9592 EESKRPVPEKRAPAEEVGIEEPPPTKVAERHMKITQEEKVLVAVTK-KEAPPRARVPEEP 9650
Query 98 KQLEEARRLAALQKKRELKAAGIITGAKRRTRKNFQPYEEVPFEEKPPPGFYEVPSEE 155
K++ R L+ +RE + +T ++R K + EVP E P V E+
Sbjct 9651 KKVAPEERFPKLKPRREEEPPAKVTEVRKRAVKEEKVSIEVPKREPRPTKEVTVTEEK 9708
> pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog
A; K13849 reticulocyte-binding protein
Length=3130
Score = 30.8 bits (68), Expect = 3.5, Method: Composition-based stats.
Identities = 34/140 (24%), Positives = 65/140 (46%), Gaps = 28/140 (20%)
Query 70 EDEKEMLEEARARLANTRGKKAKRKAREKQLEEARRLAALQKKRELKAAGIITGAKRRTR 129
+ E+E+ + + RL + ++ +++ K+ E+ R LQK+ LK R+ +
Sbjct 2748 QKEEELKRQEQERLEREKQEQLQKEEELKRQEQER----LQKEEALK---------RQEQ 2794
Query 130 KNFQPYEEVPFEEKPPPGFYEVPSEENPEGNLNFANISLQHMEGSMRAREEEKLRKEDAR 189
+ Q EE+ +E+ E E E LQ E ++ +E+E+L+KE+A
Sbjct 2795 ERLQKEEELKRQEQ------ERLEREKQE--------QLQKEE-ELKRQEQERLQKEEAL 2839
Query 190 KLKRLREDHLDEYLKIHEEK 209
K + +E LK E++
Sbjct 2840 KRQEQERLQKEEELKRQEQE 2859
Lambda K H
0.313 0.133 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6623499460
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40