bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1813_orf3
Length=194
Score E
Sequences producing significant alignments: (Bits) Value
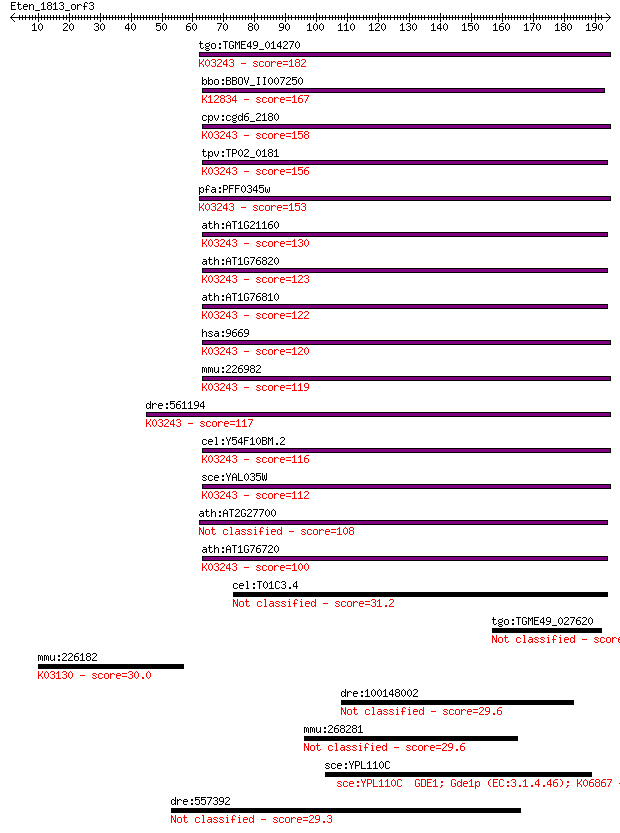
tgo:TGME49_014270 translation initiation factor IF-2, putative... 182 6e-46
bbo:BBOV_II007250 18.m06603; translation initiation factor IF-... 167 1e-41
cpv:cgd6_2180 Fun12p GTpase; translation initiation factor IF2... 158 9e-39
tpv:TP02_0181 translation initiation factor IF-2; K03243 trans... 156 4e-38
pfa:PFF0345w translation initiation factor IF-2, putative; K03... 153 4e-37
ath:AT1G21160 GTP binding / GTPase/ translation initiation fac... 130 2e-30
ath:AT1G76820 GTP binding / GTPase; K03243 translation initiat... 123 4e-28
ath:AT1G76810 eukaryotic translation initiation factor 2 famil... 122 8e-28
hsa:9669 EIF5B, DKFZp434I036, FLJ10524, IF2, KIAA0741; eukaryo... 120 3e-27
mmu:226982 Eif5b, A030003E17Rik, BC018347, IF2; eukaryotic tra... 119 6e-27
dre:561194 im:6912504; si:ch211-254d18.3; K03243 translation i... 117 3e-26
cel:Y54F10BM.2 iffb-1; Initiation Factor Five B (eIF5B) family... 116 6e-26
sce:YAL035W FUN12, yIF2; Fun12p; K03243 translation initiation... 112 7e-25
ath:AT2G27700 eukaryotic translation initiation factor 2 famil... 108 1e-23
ath:AT1G76720 GTP binding / GTPase/ translation initiation fac... 100 2e-21
cel:T01C3.4 lipase; hypothetical protein 31.2 2.3
tgo:TGME49_027620 28 kDa antigen 30.4 3.6
mmu:226182 Taf5, 6330528C20Rik, AV117817; TAF5 RNA polymerase ... 30.0 5.1
dre:100148002 pclo, wu:fj53e09; si:dkey-108d22.4 29.6 6.6
mmu:268281 Shprh, 2610103K11Rik, AA450458, AU024614, BC006883,... 29.6 7.4
sce:YPL110C GDE1; Gde1p (EC:3.1.4.46); K06867 29.6 7.5
dre:557392 MGC136778, wu:fk35e05; zgc:136778 29.3 8.3
> tgo:TGME49_014270 translation initiation factor IF-2, putative
(EC:2.7.7.4); K03243 translation initiation factor 5B
Length=1144
Score = 182 bits (462), Expect = 6e-46, Method: Compositional matrix adjust.
Identities = 84/133 (63%), Positives = 113/133 (84%), Gaps = 0/133 (0%)
Query 62 EEMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLV 121
+E+RVKG+Y H+F++AAMGVKI AP LE+AVAGTS+FV E+++DI +L+ EVM ++ +
Sbjct 837 KELRVKGEYIQHAFIQAAMGVKISAPNLEEAVAGTSVFVVEEDDDIEDLKEEVMSDMGSI 896
Query 122 LKKVEKTTNGVYVMASTLGALEALLAYLEDCKIPVFAVNIGTVQKKDIKKASVMREKGFA 181
K V++T +GVYVMASTLG+LEALL +L++ KIPVF VNIGTVQKKD+KKAS+MREKG
Sbjct 897 FKSVDRTGSGVYVMASTLGSLEALLVFLQESKIPVFGVNIGTVQKKDVKKASIMREKGRP 956
Query 182 EFAVVLGFDVKVD 194
+ +V+L F+VKVD
Sbjct 957 DLSVILAFNVKVD 969
> bbo:BBOV_II007250 18.m06603; translation initiation factor IF-2;
K12834 PHD finger-like domain-containing protein 5A
Length=1033
Score = 167 bits (424), Expect = 1e-41, Method: Composition-based stats.
Identities = 81/130 (62%), Positives = 106/130 (81%), Gaps = 0/130 (0%)
Query 63 EMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLVL 122
E+RVKG+Y H+ ++AAMGVK+VA GLE+ VAGT L + E ++DI +L +VMQ++S +
Sbjct 726 ELRVKGEYVKHTSIKAAMGVKLVAQGLEETVAGTELLLVEDDDDIEQLCEDVMQDMSSIF 785
Query 123 KKVEKTTNGVYVMASTLGALEALLAYLEDCKIPVFAVNIGTVQKKDIKKASVMREKGFAE 182
V +T GVYVMASTLG+LEALL +L D KIPVF+VNIGTVQKKD+KKAS+MREKG+ E
Sbjct 786 GNVNRTGVGVYVMASTLGSLEALLQFLTDKKIPVFSVNIGTVQKKDVKKASIMREKGYPE 845
Query 183 FAVVLGFDVK 192
++V+L FDVK
Sbjct 846 YSVILAFDVK 855
> cpv:cgd6_2180 Fun12p GTpase; translation initiation factor IF2
; K03243 translation initiation factor 5B
Length=896
Score = 158 bits (400), Expect = 9e-39, Method: Composition-based stats.
Identities = 78/135 (57%), Positives = 102/135 (75%), Gaps = 3/135 (2%)
Query 63 EMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKE---EDIAELESEVMQEVS 119
EMRVKG+Y H F++A+MGVKI A GL+DAVAGT L V K E+I L+ EVM+++
Sbjct 584 EMRVKGEYIHHRFIKASMGVKICANGLDDAVAGTQLLVQSKNSTPEEIESLKEEVMKDMG 643
Query 120 LVLKKVEKTTNGVYVMASTLGALEALLAYLEDCKIPVFAVNIGTVQKKDIKKASVMREKG 179
+ V++T NGVYVMASTLG+LEALL +L+ IPV A+NIGTV K D+++AS+M E+G
Sbjct 644 DIFSSVDRTGNGVYVMASTLGSLEALLVFLKSSNIPVVALNIGTVHKSDVRRASIMHERG 703
Query 180 FAEFAVVLGFDVKVD 194
F E AV+L FD+KVD
Sbjct 704 FPEMAVILAFDIKVD 718
> tpv:TP02_0181 translation initiation factor IF-2; K03243 translation
initiation factor 5B
Length=921
Score = 156 bits (395), Expect = 4e-38, Method: Composition-based stats.
Identities = 77/131 (58%), Positives = 103/131 (78%), Gaps = 0/131 (0%)
Query 63 EMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLVL 122
E+RVKG+Y HS ++AAM VKIVA GL+D VAGT LFV + +D+ EL +EVM ++S +
Sbjct 614 ELRVKGEYVKHSHIKAAMSVKIVANGLDDTVAGTELFVVGEGDDVDELCTEVMSDISSIF 673
Query 123 KKVEKTTNGVYVMASTLGALEALLAYLEDCKIPVFAVNIGTVQKKDIKKASVMREKGFAE 182
+++T GVYVMASTLG+LEALL +L D KI +++VNIG VQKKD+KKAS+MREKG E
Sbjct 674 DCIDRTGIGVYVMASTLGSLEALLHFLNDKKIKIYSVNIGPVQKKDVKKASIMREKGHPE 733
Query 183 FAVVLGFDVKV 193
++ +L FD+KV
Sbjct 734 YSTILAFDIKV 744
> pfa:PFF0345w translation initiation factor IF-2, putative; K03243
translation initiation factor 5B
Length=977
Score = 153 bits (386), Expect = 4e-37, Method: Composition-based stats.
Identities = 77/133 (57%), Positives = 101/133 (75%), Gaps = 0/133 (0%)
Query 62 EEMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLV 121
+E+R+K +Y H +++A +GVKI A LE+ + GTSLFV E+ E + +VM +VS V
Sbjct 671 KELRIKNEYIHHKYIKACIGVKISANNLEEVLCGTSLFVVNNIEEEEEYKKKVMTDVSDV 730
Query 122 LKKVEKTTNGVYVMASTLGALEALLAYLEDCKIPVFAVNIGTVQKKDIKKASVMREKGFA 181
V+KT G+YVMASTLG+LEALL +L D KIPVF VNIGT+QKKD+KKAS+MREKG
Sbjct 731 FNHVDKTGVGLYVMASTLGSLEALLIFLNDSKIPVFGVNIGTIQKKDVKKASIMREKGRP 790
Query 182 EFAVVLGFDVKVD 194
E+AV+L FDVK+D
Sbjct 791 EYAVILAFDVKID 803
> ath:AT1G21160 GTP binding / GTPase/ translation initiation factor;
K03243 translation initiation factor 5B
Length=1092
Score = 130 bits (328), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 70/133 (52%), Positives = 90/133 (67%), Gaps = 3/133 (2%)
Query 63 EMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLVL 122
EMRV G Y H V+AA G+KI A GLE A+AGT+L V ED+ E + M+++ V+
Sbjct 776 EMRVTGTYMPHREVKAAQGIKIAAQGLEHAIAGTALHVIGPNEDMEEAKKNAMEDIESVM 835
Query 123 KKVEKTTNGVYVMASTLGALEALLAYLE--DCKIPVFAVNIGTVQKKDIKKASVMREKGF 180
+++K+ GVYV ASTLG+LEALL +L+ D KIPV + IG V KKDI KA VM EK
Sbjct 836 NRIDKSGEGVYVQASTLGSLEALLEFLKSSDVKIPVSGIGIGPVHKKDIMKAGVMLEKK- 894
Query 181 AEFAVVLGFDVKV 193
EFA +L FDVK+
Sbjct 895 KEFATILAFDVKI 907
> ath:AT1G76820 GTP binding / GTPase; K03243 translation initiation
factor 5B
Length=1166
Score = 123 bits (308), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 67/133 (50%), Positives = 89/133 (66%), Gaps = 3/133 (2%)
Query 63 EMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLVL 122
E+RVKG Y H ++AA G+KI A GLE A+AGTSL V ++DI ++ M+++ VL
Sbjct 856 ELRVKGTYLHHKEIKAAQGIKITAQGLEHAIAGTSLHVVGPDDDIEAMKESAMEDMESVL 915
Query 123 KKVEKTTNGVYVMASTLGALEALLAYLED--CKIPVFAVNIGTVQKKDIKKASVMREKGF 180
+++K+ GVYV STLG+LEALL +L+ IPV + IG V KKDI KA VM EK
Sbjct 916 SRIDKSGEGVYVQTSTLGSLEALLEFLKTPAVNIPVSGIGIGPVHKKDIMKAGVMLEKK- 974
Query 181 AEFAVVLGFDVKV 193
E+A +L FDVKV
Sbjct 975 KEYATILAFDVKV 987
> ath:AT1G76810 eukaryotic translation initiation factor 2 family
protein / eIF-2 family protein; K03243 translation initiation
factor 5B
Length=1294
Score = 122 bits (306), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 66/133 (49%), Positives = 91/133 (68%), Gaps = 3/133 (2%)
Query 63 EMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLVL 122
E+RVKG Y + ++AA G+KI A GLE A+AGT+L V ++DI ++ M+++ VL
Sbjct 984 ELRVKGTYLHYKEIKAAQGIKITAQGLEHAIAGTALHVVGPDDDIEAIKESAMEDMESVL 1043
Query 123 KKVEKTTNGVYVMASTLGALEALLAYLED--CKIPVFAVNIGTVQKKDIKKASVMREKGF 180
+++K+ GVYV ASTLG+LEALL YL+ KIPV + IG V KKD+ KA VM E+
Sbjct 1044 SRIDKSGEGVYVQASTLGSLEALLEYLKSPAVKIPVSGIGIGPVHKKDVMKAGVMLERK- 1102
Query 181 AEFAVVLGFDVKV 193
E+A +L FDVKV
Sbjct 1103 KEYATILAFDVKV 1115
> hsa:9669 EIF5B, DKFZp434I036, FLJ10524, IF2, KIAA0741; eukaryotic
translation initiation factor 5B; K03243 translation initiation
factor 5B
Length=1220
Score = 120 bits (301), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 63/132 (47%), Positives = 88/132 (66%), Gaps = 1/132 (0%)
Query 63 EMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLVL 122
E+RVK Y+ H VEAA GVKI+ LE +AG L VA KE++I L+ E++ E+ L
Sbjct 912 ELRVKNQYEKHKEVEAAQGVKILGKDLEKTLAGLPLLVAYKEDEIPVLKDELIHELKQTL 971
Query 123 KKVEKTTNGVYVMASTLGALEALLAYLEDCKIPVFAVNIGTVQKKDIKKASVMREKGFAE 182
++ GVYV ASTLG+LEALL +L+ ++P +NIG V KKD+ KASVM E +
Sbjct 972 NAIKLEEKGVYVQASTLGSLEALLEFLKTSEVPYAGINIGPVHKKDVMKASVMLEHD-PQ 1030
Query 183 FAVVLGFDVKVD 194
+AV+L FDV+++
Sbjct 1031 YAVILAFDVRIE 1042
> mmu:226982 Eif5b, A030003E17Rik, BC018347, IF2; eukaryotic translation
initiation factor 5B; K03243 translation initiation
factor 5B
Length=1216
Score = 119 bits (298), Expect = 6e-27, Method: Compositional matrix adjust.
Identities = 62/132 (46%), Positives = 88/132 (66%), Gaps = 1/132 (0%)
Query 63 EMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLVL 122
E+RVK Y+ H VEAA GVKI+ LE +AG L VA K+++I L+ E++ E+ L
Sbjct 908 ELRVKNQYEKHKEVEAAQGVKILGKDLEKTLAGLPLLVAYKDDEIPVLKDELIHELKQTL 967
Query 123 KKVEKTTNGVYVMASTLGALEALLAYLEDCKIPVFAVNIGTVQKKDIKKASVMREKGFAE 182
++ GVYV ASTLG+LEALL +L+ ++P +NIG V KKD+ KASVM E +
Sbjct 968 NAIKLEEKGVYVQASTLGSLEALLEFLKTSEVPYAGINIGPVHKKDVMKASVMLEHD-PQ 1026
Query 183 FAVVLGFDVKVD 194
+AV+L FDV+++
Sbjct 1027 YAVILAFDVRIE 1038
> dre:561194 im:6912504; si:ch211-254d18.3; K03243 translation
initiation factor 5B
Length=1166
Score = 117 bits (292), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 64/150 (42%), Positives = 90/150 (60%), Gaps = 4/150 (2%)
Query 45 LVVSVRGDSCSESYSYSEEMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKE 104
+V +RG +E+RVK Y+ H V A GVKI+ LE +AG L VA KE
Sbjct 843 IVTQIRG---LLLPPPLKELRVKNQYEKHKEVCTAQGVKILGKDLEKTLAGLPLLVAHKE 899
Query 105 EDIAELESEVMQEVSLVLKKVEKTTNGVYVMASTLGALEALLAYLEDCKIPVFAVNIGTV 164
+++ L E+++E+ L ++ GVYV ASTLG+LEALL +L K+P +NIG V
Sbjct 900 DEVPVLRDELVRELKQTLNAIKLEEKGVYVQASTLGSLEALLEFLRTSKVPYAGINIGPV 959
Query 165 QKKDIKKASVMREKGFAEFAVVLGFDVKVD 194
KKD+ KAS M E ++AV+L FDVK++
Sbjct 960 HKKDVMKASTMLEHD-PQYAVILAFDVKIE 988
> cel:Y54F10BM.2 iffb-1; Initiation Factor Five B (eIF5B) family
member (iffb-1); K03243 translation initiation factor 5B
Length=1074
Score = 116 bits (290), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 60/132 (45%), Positives = 88/132 (66%), Gaps = 1/132 (0%)
Query 63 EMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLVL 122
E+RVK +Y + V+ A GVK++A LE +AG +++ ++E+++ L E ++++ L
Sbjct 766 ELRVKNEYIHYKEVKGARGVKVLAKNLEKVLAGLPIYITDREDEVDYLRHEADRQLANAL 825
Query 123 KKVEKTTNGVYVMASTLGALEALLAYLEDCKIPVFAVNIGTVQKKDIKKASVMREKGFAE 182
+ K GVYV ASTLG+LEALL +L+ IP VNIG V KKD++KAS M+E AE
Sbjct 826 HAIRKKPEGVYVQASTLGSLEALLEFLKSQNIPYSNVNIGPVHKKDVQKASAMKEHK-AE 884
Query 183 FAVVLGFDVKVD 194
+A VL FDVKV+
Sbjct 885 YACVLAFDVKVE 896
> sce:YAL035W FUN12, yIF2; Fun12p; K03243 translation initiation
factor 5B
Length=1002
Score = 112 bits (280), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 68/132 (51%), Positives = 92/132 (69%), Gaps = 1/132 (0%)
Query 63 EMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLVL 122
E+R+K +Y H V+AA+GVKI A LE AV+G+ L V E+D EL +VM +++ +L
Sbjct 686 ELRLKSEYVHHKEVKAALGVKIAANDLEKAVSGSRLLVVGPEDDEDELMDDVMDDLTGLL 745
Query 123 KKVEKTTNGVYVMASTLGALEALLAYLEDCKIPVFAVNIGTVQKKDIKKASVMREKGFAE 182
V+ T GV V ASTLG+LEALL +L+D KIPV ++ +G V K+D+ KAS M EK E
Sbjct 746 DSVDTTGKGVVVQASTLGSLEALLDFLKDMKIPVMSIGLGPVYKRDVMKASTMLEKA-PE 804
Query 183 FAVVLGFDVKVD 194
+AV+L FDVKVD
Sbjct 805 YAVMLCFDVKVD 816
> ath:AT2G27700 eukaryotic translation initiation factor 2 family
protein / eIF-2 family protein
Length=480
Score = 108 bits (269), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 58/134 (43%), Positives = 89/134 (66%), Gaps = 3/134 (2%)
Query 62 EEMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLV 121
+E+ V G++ H ++AA + I+A LE + GT+L V ++DI ++ VM++V+ V
Sbjct 315 KELHVNGNHVHHEVIKAAECINIIAKDLEHVIVGTALHVVGPDDDIEAIKELVMEDVNSV 374
Query 122 LKKVEKTTNGVYVMASTLGALEALLAYLED--CKIPVFAVNIGTVQKKDIKKASVMREKG 179
L +++K+ GVY+ ASTLG+LEALL +L+ K+PV + IG VQKKD+ KA VM E+
Sbjct 375 LSRIDKSGEGVYIQASTLGSLEALLEFLKSPAVKLPVGGIGIGPVQKKDVMKAGVMLERK 434
Query 180 FAEFAVVLGFDVKV 193
EFA +L DV+V
Sbjct 435 -KEFATILALDVEV 447
> ath:AT1G76720 GTP binding / GTPase/ translation initiation factor;
K03243 translation initiation factor 5B
Length=1191
Score = 100 bits (250), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 59/131 (45%), Positives = 82/131 (62%), Gaps = 8/131 (6%)
Query 63 EMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLVL 122
E+RVKG Y H ++AA G+KI A GLE A+AGTSL V ++DI ++ M+++ VL
Sbjct 907 ELRVKGTYLHHKEIKAAQGIKITAQGLEHAIAGTSLHVVGPDDDIEAMKESAMEDMESVL 966
Query 123 KKVEKTTNGVYVMASTLGALEALLAYLEDCKIPVFAVNIGTVQKKDIKKASVMREKGFAE 182
+++K+ GVYV STLG+LEALL +L K P AVNI + + + EK E
Sbjct 967 SRIDKSGEGVYVQTSTLGSLEALLEFL---KTP--AVNIPVSEGYNEGWSDAREEK---E 1018
Query 183 FAVVLGFDVKV 193
+A +L FDVKV
Sbjct 1019 YATILAFDVKV 1029
> cel:T01C3.4 lipase; hypothetical protein
Length=329
Score = 31.2 bits (69), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 54/122 (44%), Gaps = 7/122 (5%)
Query 73 HSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLVLKKVEKTTNGV 132
H +AA+ V A G +++ + E +AEL + Q+ + L +T N
Sbjct 74 HGNSDAALHVSSAATGWSNSIE----YFLEHGYTVAELYATSWQDTN-ALHAASRTHN-C 127
Query 133 YVMASTLGALEALLAYLEDCKIPVFAVNIG-TVQKKDIKKASVMREKGFAEFAVVLGFDV 191
+ LEA+LAY KI V ++G T+ +K IK SV G + + L V
Sbjct 128 KDLVRLRKFLEAVLAYTAAPKISVVTHSMGVTLARKIIKGGSVSASDGSCDLGLPLNKKV 187
Query 192 KV 193
+V
Sbjct 188 EV 189
> tgo:TGME49_027620 28 kDa antigen
Length=185
Score = 30.4 bits (67), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 18/37 (48%), Positives = 22/37 (59%), Gaps = 2/37 (5%)
Query 157 FAVNIGTVQKKDIKKASVMREKGF--AEFAVVLGFDV 191
FA N+G +K KKA V+ EKGF A+ V GF V
Sbjct 89 FAENVGQHSEKAFKKAKVVAEKGFTAAKTHTVRGFKV 125
> mmu:226182 Taf5, 6330528C20Rik, AV117817; TAF5 RNA polymerase
II, TATA box binding protein (TBP)-associated factor; K03130
transcription initiation factor TFIID subunit 5
Length=801
Score = 30.0 bits (66), Expect = 5.1, Method: Composition-based stats.
Identities = 18/47 (38%), Positives = 27/47 (57%), Gaps = 0/47 (0%)
Query 10 LRLKASLVIKRLAGSGSGFAAAAAAATAAAAILLLLVVSVRGDSCSE 56
LR +A L+ + +AGSG+ A A AA+A+L + SV G + E
Sbjct 117 LRREARLLEEAVAGSGAPGELDGAGAEAASALLSRVTASVPGSAAPE 163
> dre:100148002 pclo, wu:fj53e09; si:dkey-108d22.4
Length=3517
Score = 29.6 bits (65), Expect = 6.6, Method: Composition-based stats.
Identities = 26/84 (30%), Positives = 42/84 (50%), Gaps = 11/84 (13%)
Query 108 AELESEVMQEVSLVLKKVEKTTNGVYVMASTLGALEALLAY-----LEDCKIPVFAVNIG 162
AE +++VM E+ + LKK E T G +++ L + L D + ++ VN+
Sbjct 3375 AEGKTQVMGEIKIALKK-EMKTEGEHLVLEILQCRNITYKFKSPDHLPDLYVKLYVVNVA 3433
Query 163 TVQKKDIKKASVM----REKGFAE 182
T QK+ IKK + + RE F E
Sbjct 3434 T-QKRIIKKKTRVCRHDREPSFNE 3456
> mmu:268281 Shprh, 2610103K11Rik, AA450458, AU024614, BC006883,
D230017O13Rik, E130018M05; SNF2 histone linker PHD RING helicase
Length=1674
Score = 29.6 bits (65), Expect = 7.4, Method: Composition-based stats.
Identities = 18/69 (26%), Positives = 35/69 (50%), Gaps = 0/69 (0%)
Query 96 TSLFVAEKEEDIAELESEVMQEVSLVLKKVEKTTNGVYVMASTLGALEALLAYLEDCKIP 155
+ LF + A E + E LV +V TT G++ ++ T +++A+L++ +
Sbjct 1246 SKLFFNTVKGQTAIFEEMIEDEEGLVDDRVPTTTRGLWAVSETERSMKAILSFARSHRFD 1305
Query 156 VFAVNIGTV 164
V V+ G+V
Sbjct 1306 VEYVDEGSV 1314
> sce:YPL110C GDE1; Gde1p (EC:3.1.4.46); K06867
Length=1223
Score = 29.6 bits (65), Expect = 7.5, Method: Composition-based stats.
Identities = 23/87 (26%), Positives = 43/87 (49%), Gaps = 1/87 (1%)
Query 103 KEEDIAELESEVMQEVSLVLKKVEKTTNGVYVMASTLGALEALLAYLEDCKIPV-FAVNI 161
+EE++ ++ E+ V VLK V NG ++ S+ ++ L+ IP+ F
Sbjct 1070 EEEELGQIMMEMNHWVDTVLKVVFDNANGRDIIFSSFHPDICIMLSLKQPVIPILFLTEG 1129
Query 162 GTVQKKDIKKASVMREKGFAEFAVVLG 188
G+ Q D++ +S+ FA+ +LG
Sbjct 1130 GSEQMADLRASSLQNGIRFAKKWNLLG 1156
> dre:557392 MGC136778, wu:fk35e05; zgc:136778
Length=460
Score = 29.3 bits (64), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 29/118 (24%), Positives = 52/118 (44%), Gaps = 12/118 (10%)
Query 53 SCSESYSYSEEMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGT----SLFVAEKEE-DI 107
S E ++EEM+VK F + +K A E V F+ ++EE I
Sbjct 144 SLQEKLKHNEEMKVK-------FEKTVQHIKSRADHTERQVKHEFEKLHQFLRDEEEATI 196
Query 108 AELESEVMQEVSLVLKKVEKTTNGVYVMASTLGALEALLAYLEDCKIPVFAVNIGTVQ 165
L E Q+ ++ +K+E+ + ++ T+ E +L + C + F V++ VQ
Sbjct 197 TALREEEEQKKQMMKEKLEEMNTHISALSHTIKDTEEMLKANDVCFLKEFPVSMERVQ 254
Lambda K H
0.317 0.131 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5608917492
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40