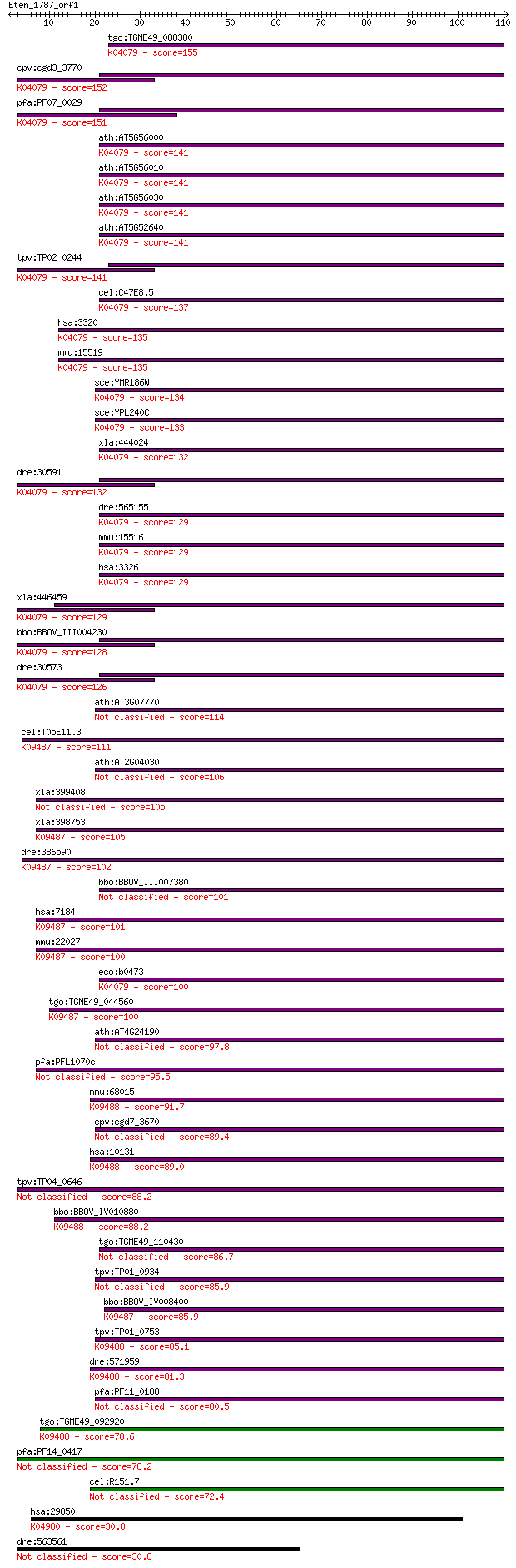
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1787_orf1
Length=110
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_088380 heat shock protein 90 ; K04079 molecular cha... 155 2e-38
cpv:cgd3_3770 Hsp90 ; K04079 molecular chaperone HtpG 152 2e-37
pfa:PF07_0029 heat shock protein 86; K04079 molecular chaperon... 151 4e-37
ath:AT5G56000 heat shock protein 81-4 (HSP81-4); K04079 molecu... 141 4e-34
ath:AT5G56010 HSP81-3; ATP binding / unfolded protein binding;... 141 4e-34
ath:AT5G56030 HSP81-2 (HEAT SHOCK PROTEIN 81-2); ATP binding; ... 141 4e-34
ath:AT5G52640 ATHSP90.1 (HEAT SHOCK PROTEIN 90.1); ATP binding... 141 5e-34
tpv:TP02_0244 heat shock protein 90; K04079 molecular chaperon... 141 6e-34
cel:C47E8.5 daf-21; abnormal DAuer Formation family member (da... 137 1e-32
hsa:3320 HSP90AA1, FLJ31884, HSP86, HSP89A, HSP90A, HSP90N, HS... 135 3e-32
mmu:15519 Hsp90aa1, 86kDa, 89kDa, AL024080, AL024147, Hsp86-1,... 135 3e-32
sce:YMR186W HSC82, HSP90; Cytoplasmic chaperone of the Hsp90 f... 134 6e-32
sce:YPL240C HSP82, HSP90; Hsp82p; K04079 molecular chaperone HtpG 133 1e-31
xla:444024 hsp90aa1.1, MGC82579, hsp86, hsp89, hsp90, hsp90a, ... 132 4e-31
dre:30591 hsp90a.1, fb17b01, hsp90, hsp90a, hsp90alpha, wu:fb1... 132 4e-31
dre:565155 hsp90a.2, cb820, hsp90a2; heat shock protein 90-alp... 129 2e-30
mmu:15516 Hsp90ab1, 90kDa, AL022974, C81438, Hsp84, Hsp84-1, H... 129 2e-30
hsa:3326 HSP90AB1, D6S182, FLJ26984, HSP90-BETA, HSP90B, HSPC2... 129 2e-30
xla:446459 hsp90ab1, hsp90b, hsp90beta; heat shock protein 90k... 129 2e-30
bbo:BBOV_III004230 17.m07381; hsp90 protein; K04079 molecular ... 128 4e-30
dre:30573 hsp90ab1, hsp90b, hsp90beta, wu:fa29f01, wu:fa91e11,... 126 2e-29
ath:AT3G07770 ATP binding 114 6e-26
cel:T05E11.3 hypothetical protein; K09487 heat shock protein 9... 111 5e-25
ath:AT2G04030 CR88; CR88; ATP binding 106 2e-23
xla:399408 hsp90b1, ecgp, gp96, grp94, tra1; heat shock protei... 105 2e-23
xla:398753 hypothetical protein MGC68448; K09487 heat shock pr... 105 5e-23
dre:386590 hsp90b1, GP96, GRP94, fb61d09, tra-1, tra1, wu:fb61... 102 3e-22
bbo:BBOV_III007380 17.m07646; heat shock protein 90 101 5e-22
hsa:7184 HSP90B1, ECGP, GP96, GRP94, TRA1; heat shock protein ... 101 7e-22
mmu:22027 Hsp90b1, ERp99, GRP94, TA-3, Targ2, Tra-1, Tra1, end... 100 8e-22
eco:b0473 htpG, ECK0467, JW0462; molecular chaperone HSP90 fam... 100 9e-22
tgo:TGME49_044560 heat shock protein 90, putative (EC:2.7.13.3... 100 1e-21
ath:AT4G24190 SHD; SHD (SHEPHERD); ATP binding / unfolded prot... 97.8 7e-21
pfa:PFL1070c endoplasmin homolog precursor, putative 95.5 4e-20
mmu:68015 Trap1, 2410002K23Rik, HSP75; TNF receptor-associated... 91.7 5e-19
cpv:cgd7_3670 heat shock protein 90 (Hsp90), signal peptide pl... 89.4 3e-18
hsa:10131 TRAP1, HSP75, HSP90L; TNF receptor-associated protei... 89.0 3e-18
tpv:TP04_0646 heat shock protein 90 88.2
bbo:BBOV_IV010880 23.m05763; heat shock protein 75; K09488 TNF... 88.2 5e-18
tgo:TGME49_110430 heat shock protein 90, putative (EC:3.2.1.3 ... 86.7 2e-17
tpv:TP01_0934 heat shock protein 90 85.9
bbo:BBOV_IV008400 23.m06066; heat shock protein 90; K09487 hea... 85.9 3e-17
tpv:TP01_0753 heat shock protein 75; K09488 TNF receptor-assoc... 85.1 4e-17
dre:571959 trap1, fc85a11, wu:fc85a11; TNF receptor-associated... 81.3 7e-16
pfa:PF11_0188 heat shock protein 90, putative 80.5 1e-15
tgo:TGME49_092920 heat shock protein 90, putative ; K09488 TNF... 78.6 5e-15
pfa:PF14_0417 HSP90 78.2 6e-15
cel:R151.7 hypothetical protein 72.4 4e-13
hsa:29850 TRPM5, LTRPC5, MTR1; transient receptor potential ca... 30.8 1.1
dre:563561 myc box-dependent-interacting protein 1-like 30.8 1.2
> tgo:TGME49_088380 heat shock protein 90 ; K04079 molecular chaperone
HtpG
Length=708
Score = 155 bits (393), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 76/87 (87%), Positives = 81/87 (93%), Gaps = 0/87 (0%)
Query 23 ADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLIPD 82
ADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAIT+PEKLK LFIR++P+
Sbjct 11 ADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITDPEKLKGAERLFIRIVPN 70
Query 83 KANNTLTIEDSGIGMTKAELVNNLGTL 109
K NNTLTIED GIGMTKAELVNNLGT+
Sbjct 71 KQNNTLTIEDDGIGMTKAELVNNLGTI 97
> cpv:cgd3_3770 Hsp90 ; K04079 molecular chaperone HtpG
Length=711
Score = 152 bits (385), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 73/89 (82%), Positives = 82/89 (92%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
ADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYE++T+PE+LK+ E+ IR+I
Sbjct 19 FNADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYESLTDPEQLKSNEEMHIRII 78
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
PDK NNTLTIEDSGIGMTK EL+NNLGT+
Sbjct 79 PDKVNNTLTIEDSGIGMTKNELINNLGTI 107
Score = 28.5 bits (62), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 3 KEGLEIDESEEEKKKFEELKADIQQLMSLI 32
KEGL ++E+ EEK+ FE L+ + + L LI
Sbjct 530 KEGLTLEETAEEKEAFEALQKEYEPLCQLI 559
> pfa:PF07_0029 heat shock protein 86; K04079 molecular chaperone
HtpG
Length=745
Score = 151 bits (382), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 72/89 (80%), Positives = 81/89 (91%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
ADI+QLMSLIINTFYSNKEIFLRELISNASDALDKIRYE+IT+ +KL +PE FIR+I
Sbjct 8 FNADIRQLMSLIINTFYSNKEIFLRELISNASDALDKIRYESITDTQKLSAEPEFFIRII 67
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
PDK NNTLTIEDSGIGMTK +L+NNLGT+
Sbjct 68 PDKTNNTLTIEDSGIGMTKNDLINNLGTI 96
Score = 36.2 bits (82), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 18/35 (51%), Positives = 25/35 (71%), Gaps = 0/35 (0%)
Query 3 KEGLEIDESEEEKKKFEELKADIQQLMSLIINTFY 37
KEGL+ID+SEE KK FE LKA+ + L +I + +
Sbjct 565 KEGLDIDDSEEAKKDFETLKAEYEGLCKVIKDVLH 599
> ath:AT5G56000 heat shock protein 81-4 (HSP81-4); K04079 molecular
chaperone HtpG
Length=699
Score = 141 bits (356), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 68/89 (76%), Positives = 80/89 (89%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
+A+I QL+SLIINTFYSNKEIFLRELISN+SDALDKIR+E++T+ KL +PELFI +I
Sbjct 9 FQAEINQLLSLIINTFYSNKEIFLRELISNSSDALDKIRFESLTDKSKLDGQPELFIHII 68
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
PDK NNTLTI DSGIGMTKA+LVNNLGT+
Sbjct 69 PDKTNNTLTIIDSGIGMTKADLVNNLGTI 97
> ath:AT5G56010 HSP81-3; ATP binding / unfolded protein binding;
K04079 molecular chaperone HtpG
Length=699
Score = 141 bits (356), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 68/89 (76%), Positives = 80/89 (89%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
+A+I QL+SLIINTFYSNKEIFLRELISN+SDALDKIR+E++T+ KL +PELFI +I
Sbjct 9 FQAEINQLLSLIINTFYSNKEIFLRELISNSSDALDKIRFESLTDKSKLDGQPELFIHII 68
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
PDK NNTLTI DSGIGMTKA+LVNNLGT+
Sbjct 69 PDKTNNTLTIIDSGIGMTKADLVNNLGTI 97
> ath:AT5G56030 HSP81-2 (HEAT SHOCK PROTEIN 81-2); ATP binding;
K04079 molecular chaperone HtpG
Length=699
Score = 141 bits (356), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 68/89 (76%), Positives = 80/89 (89%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
+A+I QL+SLIINTFYSNKEIFLRELISN+SDALDKIR+E++T+ KL +PELFI +I
Sbjct 9 FQAEINQLLSLIINTFYSNKEIFLRELISNSSDALDKIRFESLTDKSKLDGQPELFIHII 68
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
PDK NNTLTI DSGIGMTKA+LVNNLGT+
Sbjct 69 PDKTNNTLTIIDSGIGMTKADLVNNLGTI 97
> ath:AT5G52640 ATHSP90.1 (HEAT SHOCK PROTEIN 90.1); ATP binding
/ unfolded protein binding; K04079 molecular chaperone HtpG
Length=705
Score = 141 bits (356), Expect = 5e-34, Method: Compositional matrix adjust.
Identities = 67/89 (75%), Positives = 81/89 (91%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
+A+I QL+SLIINTFYSNKEIFLRELISN+SDALDKIR+E++T+ KL +PELFIRL+
Sbjct 14 FQAEINQLLSLIINTFYSNKEIFLRELISNSSDALDKIRFESLTDKSKLDGQPELFIRLV 73
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
PDK+N TL+I DSGIGMTKA+LVNNLGT+
Sbjct 74 PDKSNKTLSIIDSGIGMTKADLVNNLGTI 102
> tpv:TP02_0244 heat shock protein 90; K04079 molecular chaperone
HtpG
Length=721
Score = 141 bits (355), Expect = 6e-34, Method: Compositional matrix adjust.
Identities = 67/87 (77%), Positives = 79/87 (90%), Gaps = 0/87 (0%)
Query 23 ADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLIPD 82
ADI QL+SLIIN FYSNKEIFLRELISNASDAL+KIRYEAI +P++++ +P+ +IRL D
Sbjct 17 ADISQLLSLIINAFYSNKEIFLRELISNASDALEKIRYEAIKDPKQIEDQPDYYIRLYAD 76
Query 83 KANNTLTIEDSGIGMTKAELVNNLGTL 109
K NNTLTIEDSGIGMTKA+LVNNLGT+
Sbjct 77 KNNNTLTIEDSGIGMTKADLVNNLGTI 103
Score = 32.7 bits (73), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 16/30 (53%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 3 KEGLEIDESEEEKKKFEELKADIQQLMSLI 32
KEGL++DE E+EKK FE LK +++ L I
Sbjct 541 KEGLDLDEGEDEKKSFEALKEEMEPLCKHI 570
> cel:C47E8.5 daf-21; abnormal DAuer Formation family member (daf-21);
K04079 molecular chaperone HtpG
Length=702
Score = 137 bits (344), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 66/89 (74%), Positives = 78/89 (87%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
+A+I QLMSLIINTFYSNKEI+LRELISNASDALDKIRY+A+TEP +L T ELFI++
Sbjct 10 FQAEIAQLMSLIINTFYSNKEIYLRELISNASDALDKIRYQALTEPSELDTGKELFIKIT 69
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
P+K TLTI D+GIGMTKA+LVNNLGT+
Sbjct 70 PNKEEKTLTIMDTGIGMTKADLVNNLGTI 98
> hsa:3320 HSP90AA1, FLJ31884, HSP86, HSP89A, HSP90A, HSP90N,
HSPC1, HSPCA, HSPCAL1, HSPCAL4, HSPN, Hsp89, Hsp90, LAP2; heat
shock protein 90kDa alpha (cytosolic), class A member 1;
K04079 molecular chaperone HtpG
Length=854
Score = 135 bits (341), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 69/98 (70%), Positives = 83/98 (84%), Gaps = 1/98 (1%)
Query 12 EEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKT 71
EEE + F +A+I QLMSLIINTFYSNKEIFLRELISN+SDALDKIRYE++T+P KL +
Sbjct 136 EEEVETFA-FQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYESLTDPSKLDS 194
Query 72 KPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
EL I LIP+K + TLTI D+GIGMTKA+L+NNLGT+
Sbjct 195 GKELHINLIPNKQDRTLTIVDTGIGMTKADLINNLGTI 232
> mmu:15519 Hsp90aa1, 86kDa, 89kDa, AL024080, AL024147, Hsp86-1,
Hsp89, Hsp90, Hspca, hsp4; heat shock protein 90, alpha (cytosolic),
class A member 1; K04079 molecular chaperone HtpG
Length=733
Score = 135 bits (340), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 69/98 (70%), Positives = 82/98 (83%), Gaps = 1/98 (1%)
Query 12 EEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKT 71
EEE + F +A+I QLMSLIINTFYSNKEIFLRELISN+SDALDKIRYE++T+P KL +
Sbjct 14 EEEVETFA-FQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYESLTDPSKLDS 72
Query 72 KPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
EL I LIP K + TLTI D+GIGMTKA+L+NNLGT+
Sbjct 73 GKELHINLIPSKQDRTLTIVDTGIGMTKADLINNLGTI 110
> sce:YMR186W HSC82, HSP90; Cytoplasmic chaperone of the Hsp90
family, redundant in function and nearly identical with Hsp82p,
and together they are essential; expressed constitutively
at 10-fold higher basal levels than HSP82 and induced 2-3
fold by heat shock; K04079 molecular chaperone HtpG
Length=705
Score = 134 bits (338), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 64/90 (71%), Positives = 78/90 (86%), Gaps = 0/90 (0%)
Query 20 ELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRL 79
E +A+I QLMSLIINT YSNKEIFLRELISNASDALDKIRY+A+++P++L+T+P+LFIR+
Sbjct 7 EFQAEITQLMSLIINTVYSNKEIFLRELISNASDALDKIRYQALSDPKQLETEPDLFIRI 66
Query 80 IPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
P L I DSGIGMTKAEL+NNLGT+
Sbjct 67 TPKPEEKVLEIRDSGIGMTKAELINNLGTI 96
> sce:YPL240C HSP82, HSP90; Hsp82p; K04079 molecular chaperone
HtpG
Length=709
Score = 133 bits (335), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 63/90 (70%), Positives = 78/90 (86%), Gaps = 0/90 (0%)
Query 20 ELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRL 79
E +A+I QLMSLIINT YSNKEIFLRELISNASDALDKIRY+++++P++L+T+P+LFIR+
Sbjct 7 EFQAEITQLMSLIINTVYSNKEIFLRELISNASDALDKIRYKSLSDPKQLETEPDLFIRI 66
Query 80 IPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
P L I DSGIGMTKAEL+NNLGT+
Sbjct 67 TPKPEQKVLEIRDSGIGMTKAELINNLGTI 96
> xla:444024 hsp90aa1.1, MGC82579, hsp86, hsp89, hsp90, hsp90a,
hspc1, hspca, hspn, lap2; heat shock protein 90kDa alpha (cytosolic),
class A member 1, gene 1; K04079 molecular chaperone
HtpG
Length=729
Score = 132 bits (331), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 64/89 (71%), Positives = 78/89 (87%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
+A+I QLMSLIINTFYSNKEIFLRELISN+SDALDKIRYE++T+P KL + EL I LI
Sbjct 23 FQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYESLTDPSKLDSGKELKIELI 82
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
P+K + +LTI D+GIGMTKA+L+NNLGT+
Sbjct 83 PNKQDRSLTIIDTGIGMTKADLINNLGTI 111
> dre:30591 hsp90a.1, fb17b01, hsp90, hsp90a, hsp90alpha, wu:fb17b01,
zgc:86652; heat shock protein 90-alpha 1; K04079 molecular
chaperone HtpG
Length=726
Score = 132 bits (331), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 64/89 (71%), Positives = 77/89 (86%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
+A+I QLMSLIINTFYSNKEIFLRELISN+SDALDKIRYE++T+P KL + +L I LI
Sbjct 20 FQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYESLTDPSKLDSCKDLKIELI 79
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
PD+ TLTI D+GIGMTKA+L+NNLGT+
Sbjct 80 PDQKERTLTIIDTGIGMTKADLINNLGTI 108
Score = 32.3 bits (72), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 17/30 (56%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 3 KEGLEIDESEEEKKKFEELKADIQQLMSLI 32
KEGLE+ E EEEKKK +ELKA + L ++
Sbjct 540 KEGLELPEDEEEKKKQDELKAKYENLCKIM 569
> dre:565155 hsp90a.2, cb820, hsp90a2; heat shock protein 90-alpha
2; K04079 molecular chaperone HtpG
Length=734
Score = 129 bits (325), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 63/89 (70%), Positives = 77/89 (86%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
+A+I QLMSLIINTFYSNKEIFLRELISN+SDALDKIRYE++T+P KL + +L I +I
Sbjct 20 FQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYESLTDPSKLDSGKDLKIEII 79
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
P+K TLTI D+GIGMTKA+L+NNLGT+
Sbjct 80 PNKEERTLTIIDTGIGMTKADLINNLGTI 108
> mmu:15516 Hsp90ab1, 90kDa, AL022974, C81438, Hsp84, Hsp84-1,
Hsp90, Hspcb, MGC115780; heat shock protein 90 alpha (cytosolic),
class B member 1; K04079 molecular chaperone HtpG
Length=724
Score = 129 bits (325), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 63/89 (70%), Positives = 76/89 (85%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
+A+I QLMSLIINTFYSNKEIFLRELISNASDALDKIRYE++T+P KL + EL I +I
Sbjct 17 FQAEIAQLMSLIINTFYSNKEIFLRELISNASDALDKIRYESLTDPSKLDSGKELKIDII 76
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
P+ TLT+ D+GIGMTKA+L+NNLGT+
Sbjct 77 PNPQERTLTLVDTGIGMTKADLINNLGTI 105
> hsa:3326 HSP90AB1, D6S182, FLJ26984, HSP90-BETA, HSP90B, HSPC2,
HSPCB; heat shock protein 90kDa alpha (cytosolic), class
B member 1; K04079 molecular chaperone HtpG
Length=724
Score = 129 bits (325), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 63/89 (70%), Positives = 76/89 (85%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
+A+I QLMSLIINTFYSNKEIFLRELISNASDALDKIRYE++T+P KL + EL I +I
Sbjct 17 FQAEIAQLMSLIINTFYSNKEIFLRELISNASDALDKIRYESLTDPSKLDSGKELKIDII 76
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
P+ TLT+ D+GIGMTKA+L+NNLGT+
Sbjct 77 PNPQERTLTLVDTGIGMTKADLINNLGTI 105
> xla:446459 hsp90ab1, hsp90b, hsp90beta; heat shock protein 90kDa
alpha (cytosolic), class B member 1; K04079 molecular chaperone
HtpG
Length=722
Score = 129 bits (325), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 66/99 (66%), Positives = 82/99 (82%), Gaps = 1/99 (1%)
Query 11 SEEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLK 70
EEE + F +A+I QLMSLIINTFYSNKEIFLRELISNASDALDKIRYE++T+P KL
Sbjct 8 GEEEVETFA-FQAEIAQLMSLIINTFYSNKEIFLRELISNASDALDKIRYESLTDPSKLD 66
Query 71 TKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
+ +L I +IP++ TLT+ D+GIGMTKA+L+NNLGT+
Sbjct 67 SGKDLKIDIIPNRLERTLTMIDTGIGMTKADLINNLGTI 105
Score = 29.3 bits (64), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 16/30 (53%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 3 KEGLEIDESEEEKKKFEELKADIQQLMSLI 32
KEGLE+ E EEEKK EE K + L L+
Sbjct 536 KEGLELPEDEEEKKTMEENKTKFESLCKLM 565
> bbo:BBOV_III004230 17.m07381; hsp90 protein; K04079 molecular
chaperone HtpG
Length=712
Score = 128 bits (322), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 63/89 (70%), Positives = 74/89 (83%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
ADI QL+SLIIN FYSNKEIFLRELISNASDAL+KIRYEAI +P++++ PE I L
Sbjct 11 FNADISQLLSLIINAFYSNKEIFLRELISNASDALEKIRYEAIKDPKQVEDFPEYQISLS 70
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
DK N TLTIED+GIGMTK +L+NNLGT+
Sbjct 71 ADKTNKTLTIEDTGIGMTKTDLINNLGTI 99
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 23/30 (76%), Gaps = 0/30 (0%)
Query 3 KEGLEIDESEEEKKKFEELKADIQQLMSLI 32
KE LE++++EEE+K FE L+ +++ L LI
Sbjct 533 KENLELEDTEEERKNFETLEKEMEPLCRLI 562
> dre:30573 hsp90ab1, hsp90b, hsp90beta, wu:fa29f01, wu:fa91e11,
wu:fd59e11, wu:gcd22h07; heat shock protein 90, alpha (cytosolic),
class B member 1; K04079 molecular chaperone HtpG
Length=725
Score = 126 bits (317), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 61/89 (68%), Positives = 76/89 (85%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
+A+I QLMSLIINTFYSNKEIFLREL+SNASDALDKIRYE++T+P KL + +L I +I
Sbjct 16 FQAEIAQLMSLIINTFYSNKEIFLRELVSNASDALDKIRYESLTDPTKLDSGKDLKIDII 75
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
P+ TLT+ D+GIGMTKA+L+NNLGT+
Sbjct 76 PNVQERTLTLIDTGIGMTKADLINNLGTI 104
Score = 32.0 bits (71), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 17/30 (56%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 3 KEGLEIDESEEEKKKFEELKADIQQLMSLI 32
KEGLE+ E E+EKKK EE KA + L L+
Sbjct 537 KEGLELPEDEDEKKKMEEDKAKFENLCKLM 566
> ath:AT3G07770 ATP binding
Length=799
Score = 114 bits (286), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 54/90 (60%), Positives = 71/90 (78%), Gaps = 0/90 (0%)
Query 20 ELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRL 79
E +A++ +LM LI+N+ YSNKE+FLRELISNASDALDK+RY ++T PE K P+L IR+
Sbjct 98 EYQAEVSRLMDLIVNSLYSNKEVFLRELISNASDALDKLRYLSVTNPELSKDAPDLDIRI 157
Query 80 IPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
DK N +T+ DSGIGMT+ ELV+ LGT+
Sbjct 158 YADKENGIITLTDSGIGMTRQELVDCLGTI 187
> cel:T05E11.3 hypothetical protein; K09487 heat shock protein
90kDa beta
Length=760
Score = 111 bits (278), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 54/108 (50%), Positives = 81/108 (75%), Gaps = 2/108 (1%)
Query 4 EGLEIDESEEEKKKFE--ELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYE 61
+GL + + +E + K E E +A++ ++M LIIN+ Y NKEIFLRELISNASDALDKIR
Sbjct 47 DGLSVSQIKELRSKAEKHEFQAEVNRMMKLIINSLYRNKEIFLRELISNASDALDKIRLL 106
Query 62 AITEPEKLKTKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
++T+PE+L+ E+ +++ D+ N L I D+G+GMT+ +L+NNLGT+
Sbjct 107 SLTDPEQLRETEEMSVKIKADRENRLLHITDTGVGMTRQDLINNLGTI 154
> ath:AT2G04030 CR88; CR88; ATP binding
Length=780
Score = 106 bits (264), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 49/90 (54%), Positives = 71/90 (78%), Gaps = 0/90 (0%)
Query 20 ELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRL 79
E +A++ +L+ LI+++ YS+KE+FLREL+SNASDALDK+R+ ++TEP L +L IR+
Sbjct 80 EYQAEVSRLLDLIVHSLYSHKEVFLRELVSNASDALDKLRFLSVTEPSLLGDGGDLEIRI 139
Query 80 IPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
PD N T+TI D+GIGMTK EL++ LGT+
Sbjct 140 KPDPDNGTITITDTGIGMTKEELIDCLGTI 169
> xla:399408 hsp90b1, ecgp, gp96, grp94, tra1; heat shock protein
90kDa beta (Grp94), member 1
Length=804
Score = 105 bits (263), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 57/103 (55%), Positives = 74/103 (71%), Gaps = 1/103 (0%)
Query 7 EIDESEEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEP 66
+I E E+ +KF +A++ ++M LIIN+ Y NKEIFLRELISNASDALDKIR ++T+
Sbjct 65 QIKEIREKSEKFA-FQAEVNRMMKLIINSLYKNKEIFLRELISNASDALDKIRLMSLTDD 123
Query 67 EKLKTKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
+ L EL I++ DK N L I D+GIGMTK ELV NLGT+
Sbjct 124 QALAANEELTIKIKCDKEKNMLHITDTGIGMTKEELVKNLGTI 166
> xla:398753 hypothetical protein MGC68448; K09487 heat shock
protein 90kDa beta
Length=805
Score = 105 bits (261), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 57/103 (55%), Positives = 73/103 (70%), Gaps = 1/103 (0%)
Query 7 EIDESEEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEP 66
+I E E+ +KF +A++ ++M LIIN+ Y NKEIFLRELISNASDALDKIR ++T+
Sbjct 65 QIKEIREKSEKFA-FQAEVNRMMKLIINSLYKNKEIFLRELISNASDALDKIRLMSLTDE 123
Query 67 EKLKTKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
L EL I++ DK N L I D+GIGMTK ELV NLGT+
Sbjct 124 NALAANEELTIKIKCDKEKNMLQITDTGIGMTKEELVKNLGTI 166
> dre:386590 hsp90b1, GP96, GRP94, fb61d09, tra-1, tra1, wu:fb61d09,
wu:fq25g01; heat shock protein 90, beta (grp94), member
1; K09487 heat shock protein 90kDa beta
Length=793
Score = 102 bits (255), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 57/108 (52%), Positives = 75/108 (69%), Gaps = 2/108 (1%)
Query 4 EGLEIDESEEEKKKFEE--LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYE 61
+GL + +E + K E+ +A++ ++M LIIN+ Y NKEIFLRELISNASDALDKIR
Sbjct 59 DGLNTSQLKEIRDKAEKHAFQAEVNRMMKLIINSLYKNKEIFLRELISNASDALDKIRLL 118
Query 62 AITEPEKLKTKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
++T + L EL I++ DK N L I D+GIGMTK ELV NLGT+
Sbjct 119 SLTNEDALAGNEELTIKIKSDKEKNMLHITDTGIGMTKEELVKNLGTI 166
> bbo:BBOV_III007380 17.m07646; heat shock protein 90
Length=795
Score = 101 bits (252), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 52/91 (57%), Positives = 73/91 (80%), Gaps = 5/91 (5%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELF--IR 78
+A++ ++M +I+N+ Y++K+IFLREL+SNA+DALDK R +A +P++ K E F IR
Sbjct 125 FQAEVSRVMDIIVNSLYTDKDIFLRELVSNAADALDKRRIQA--DPDE-KVPKESFGGIR 181
Query 79 LIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
+IPDK +NTLTIED GIGMTK ELV+NLGT+
Sbjct 182 IIPDKEHNTLTIEDDGIGMTKDELVHNLGTI 212
> hsa:7184 HSP90B1, ECGP, GP96, GRP94, TRA1; heat shock protein
90kDa beta (Grp94), member 1; K09487 heat shock protein 90kDa
beta
Length=803
Score = 101 bits (251), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 53/103 (51%), Positives = 73/103 (70%), Gaps = 1/103 (0%)
Query 7 EIDESEEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEP 66
+I E E+ +KF +A++ ++M LIIN+ Y NKEIFLRELISNASDALDKIR ++T+
Sbjct 65 QIRELREKSEKFA-FQAEVNRMMKLIINSLYKNKEIFLRELISNASDALDKIRLISLTDE 123
Query 67 EKLKTKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
L EL +++ DK N L + D+G+GMT+ ELV NLGT+
Sbjct 124 NALSGNEELTVKIKCDKEKNLLHVTDTGVGMTREELVKNLGTI 166
> mmu:22027 Hsp90b1, ERp99, GRP94, TA-3, Targ2, Tra-1, Tra1, endoplasmin,
gp96; heat shock protein 90, beta (Grp94), member
1; K09487 heat shock protein 90kDa beta
Length=802
Score = 100 bits (250), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 53/103 (51%), Positives = 73/103 (70%), Gaps = 1/103 (0%)
Query 7 EIDESEEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEP 66
+I E E+ +KF +A++ ++M LIIN+ Y NKEIFLRELISNASDALDKIR ++T+
Sbjct 65 QIRELREKSEKFA-FQAEVNRMMKLIINSLYKNKEIFLRELISNASDALDKIRLISLTDE 123
Query 67 EKLKTKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
L EL +++ DK N L + D+G+GMT+ ELV NLGT+
Sbjct 124 NALAGNEELTVKIKCDKEKNLLHVTDTGVGMTREELVKNLGTI 166
> eco:b0473 htpG, ECK0467, JW0462; molecular chaperone HSP90 family;
K04079 molecular chaperone HtpG
Length=624
Score = 100 bits (250), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 45/89 (50%), Positives = 70/89 (78%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
+++++QL+ L+I++ YSNKEIFLRELISNASDA DK+R+ A++ P+ + EL +R+
Sbjct 9 FQSEVKQLLHLMIHSLYSNKEIFLRELISNASDAADKLRFRALSNPDLYEGDGELRVRVS 68
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
DK TLTI D+G+GMT+ E++++LGT+
Sbjct 69 FDKDKRTLTISDNGVGMTRDEVIDHLGTI 97
> tgo:TGME49_044560 heat shock protein 90, putative (EC:2.7.13.3);
K09487 heat shock protein 90kDa beta
Length=847
Score = 100 bits (248), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 48/100 (48%), Positives = 74/100 (74%), Gaps = 0/100 (0%)
Query 10 ESEEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKL 69
E+ ++ ++ + + ++ +LM +IIN+ Y+ +E+FLRELISNA DAL+K+R+ A++ PE L
Sbjct 79 EAVQKSQESHQYQTEVSRLMDIIINSLYTQREVFLRELISNAVDALEKVRFTALSHPEVL 138
Query 70 KTKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
+ K L IR+ D TL+I DSGIGMTK +L+NNLGT+
Sbjct 139 EPKKNLDIRIEFDADAKTLSIIDSGIGMTKQDLINNLGTV 178
> ath:AT4G24190 SHD; SHD (SHEPHERD); ATP binding / unfolded protein
binding
Length=823
Score = 97.8 bits (242), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 53/94 (56%), Positives = 70/94 (74%), Gaps = 6/94 (6%)
Query 20 ELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKL----KTKPEL 75
E +A++ +LM +IIN+ YSNK+IFLRELISNASDALDKIR+ A+T+ + L K E+
Sbjct 80 EFQAEVSRLMDIIINSLYSNKDIFLRELISNASDALDKIRFLALTDKDVLGEGDTAKLEI 139
Query 76 FIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
I+L DKA L+I D GIGMTK +L+ NLGT+
Sbjct 140 QIKL--DKAKKILSIRDRGIGMTKEDLIKNLGTI 171
> pfa:PFL1070c endoplasmin homolog precursor, putative
Length=821
Score = 95.5 bits (236), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 48/105 (45%), Positives = 77/105 (73%), Gaps = 2/105 (1%)
Query 7 EIDESEEEKKKFE--ELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAIT 64
EI+E E+ + E + + ++ +LM +I+N+ Y+ KE+FLRELISNA+DAL+KIR+ +++
Sbjct 61 EIEEGEKPTESMESHQYQTEVTRLMDIIVNSLYTQKEVFLRELISNAADALEKIRFLSLS 120
Query 65 EPEKLKTKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
+ L + +L IR+ +K N L+I D+GIGMTK +L+NNLGT+
Sbjct 121 DESVLGEEKKLEIRISANKEKNILSITDTGIGMTKVDLINNLGTI 165
> mmu:68015 Trap1, 2410002K23Rik, HSP75; TNF receptor-associated
protein 1; K09488 TNF receptor-associated protein 1
Length=706
Score = 91.7 bits (226), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 44/91 (48%), Positives = 67/91 (73%), Gaps = 3/91 (3%)
Query 19 EELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIR 78
E +A+ ++L+ ++ + YS KE+F+RELISNASDAL+K+R++ + E + L PE+ I
Sbjct 90 HEFQAETKKLLDIVARSLYSEKEVFIRELISNASDALEKLRHKLVCEGQVL---PEMEIH 146
Query 79 LIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
L D T+TI+D+GIGMT+ ELV+NLGT+
Sbjct 147 LQTDAKKGTITIQDTGIGMTQEELVSNLGTI 177
> cpv:cgd7_3670 heat shock protein 90 (Hsp90), signal peptide
plus ER retention motif
Length=787
Score = 89.4 bits (220), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 42/90 (46%), Positives = 66/90 (73%), Gaps = 0/90 (0%)
Query 20 ELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRL 79
E + ++ +LM +IIN+ YS K++FLREL+SN++DAL+K R+ ++T+ L + EL IR+
Sbjct 90 EFQTEVSRLMDIIINSLYSQKDVFLRELLSNSADALEKARFISVTDDSFLGEQQELEIRV 149
Query 80 IPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
+ T+TI D+GIGMT+ +LV NLGT+
Sbjct 150 SFNNDKRTITISDTGIGMTRHDLVTNLGTV 179
> hsa:10131 TRAP1, HSP75, HSP90L; TNF receptor-associated protein
1; K09488 TNF receptor-associated protein 1
Length=704
Score = 89.0 bits (219), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 42/91 (46%), Positives = 68/91 (74%), Gaps = 3/91 (3%)
Query 19 EELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIR 78
E +A+ ++L+ ++ + YS KE+F+RELISNASDAL+K+R++ +++ + L PE+ I
Sbjct 88 HEFQAETKKLLDIVARSLYSEKEVFIRELISNASDALEKLRHKLVSDGQAL---PEMEIH 144
Query 79 LIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
L + T+TI+D+GIGMT+ ELV+NLGT+
Sbjct 145 LQTNAEKGTITIQDTGIGMTQEELVSNLGTI 175
> tpv:TP04_0646 heat shock protein 90
Length=913
Score = 88.2 bits (217), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 51/109 (46%), Positives = 77/109 (70%), Gaps = 8/109 (7%)
Query 3 KEGLEIDESEEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEA 62
+E E+ S E+ F+ A++ ++M +I+N+ Y++++IFLREL+SN++DALDK R +A
Sbjct 120 QEPPEVSLSGEQTYPFQ---AEVSRVMDIIVNSLYTDRDIFLRELVSNSADALDKRRLKA 176
Query 63 ITEPEKLKTKPELF--IRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
+PE+ K E F IR++P+K +TLTIED GIGMT EL NLGT+
Sbjct 177 --DPEE-KIPKEAFGGIRIMPNKDLSTLTIEDDGIGMTAEELKTNLGTI 222
> bbo:BBOV_IV010880 23.m05763; heat shock protein 75; K09488 TNF
receptor-associated protein 1
Length=623
Score = 88.2 bits (217), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 47/104 (45%), Positives = 75/104 (72%), Gaps = 6/104 (5%)
Query 11 SEEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLK 70
++++K E KA+IQ+L+ ++ ++ Y++KE+F+RELISNASDAL+K+R+ T+ E L
Sbjct 10 TDQQKVDTYEFKAEIQKLLQIVAHSLYTDKEVFVRELISNASDALEKLRFLEATQ-ENLA 68
Query 71 TK---PELF--IRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
T PE+ I++ + T TIED+G+GMTK E++NNLGT+
Sbjct 69 TSKVDPEVAYKIKISTNPEEKTFTIEDTGVGMTKDEIINNLGTI 112
> tgo:TGME49_110430 heat shock protein 90, putative (EC:3.2.1.3
2.7.13.3)
Length=1100
Score = 86.7 bits (213), Expect = 2e-17, Method: Composition-based stats.
Identities = 42/89 (47%), Positives = 64/89 (71%), Gaps = 4/89 (4%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
+A++++++ +I+N+ Y+++++FLRELISNA+DA DK R E K + IR+
Sbjct 250 FQAEVKRVLDIIVNSLYTDRDVFLRELISNAADASDKKRMLMEKEGRKFRGS----IRVR 305
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
D+ NTLTIED GIGM+K EL+NNLGT+
Sbjct 306 ADREKNTLTIEDDGIGMSKTELINNLGTI 334
> tpv:TP01_0934 heat shock protein 90
Length=1009
Score = 85.9 bits (211), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 43/90 (47%), Positives = 67/90 (74%), Gaps = 1/90 (1%)
Query 20 ELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRL 79
E +A++ +L+ +I+N+ YS+K+IFLREL+SN++DAL+K + A+ + K K ELF+R+
Sbjct 84 EYQAEVTRLLDIIVNSLYSSKDIFLRELVSNSADALEKYKITALQKNYKDKD-VELFVRI 142
Query 80 IPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
LTI D+G+GMTK+EL+NNLGT+
Sbjct 143 RSYPKKRLLTIWDNGVGMTKSELMNNLGTI 172
> bbo:BBOV_IV008400 23.m06066; heat shock protein 90; K09487 heat
shock protein 90kDa beta
Length=795
Score = 85.9 bits (211), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 43/88 (48%), Positives = 65/88 (73%), Gaps = 1/88 (1%)
Query 22 KADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLIP 81
+AD ++M +I+N+ YSNK++FLRELISN++DAL+K + + E + ++ EL I++
Sbjct 94 QADFARVMDIIVNSLYSNKDVFLRELISNSADALEKYKIVELRE-NRSESVDELAIKIRV 152
Query 82 DKANNTLTIEDSGIGMTKAELVNNLGTL 109
K TLTI D+G+GMTK EL+NNLGT+
Sbjct 153 SKNKRTLTILDTGVGMTKHELINNLGTI 180
> tpv:TP01_0753 heat shock protein 75; K09488 TNF receptor-associated
protein 1
Length=724
Score = 85.1 bits (209), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 46/95 (48%), Positives = 68/95 (71%), Gaps = 6/95 (6%)
Query 20 ELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKL---KTKPELF 76
+ KA+ Q+L+ ++ ++ Y++KE+F+RELISNASDAL+K+R+ T E L K P++
Sbjct 80 QFKAETQKLLQIVAHSLYTDKEVFVRELISNASDALEKLRFLESTR-EGLAASKVDPDVG 138
Query 77 --IRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
IR+ D T TIED+G+GMTK E+VNNLGT+
Sbjct 139 YKIRISVDPKTKTFTIEDTGVGMTKEEIVNNLGTI 173
> dre:571959 trap1, fc85a11, wu:fc85a11; TNF receptor-associated
protein 1; K09488 TNF receptor-associated protein 1
Length=719
Score = 81.3 bits (199), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 41/91 (45%), Positives = 61/91 (67%), Gaps = 3/91 (3%)
Query 19 EELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIR 78
E +A+ ++L+ ++ + YS KE+F+RELISN SDAL+K+R+ IT T P + I
Sbjct 103 HEFQAETKKLLDIVARSLYSEKEVFIRELISNGSDALEKLRHRMITAGG--DTAP-MEIH 159
Query 79 LIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
L D T TI+D+G+GM K +LV+NLGT+
Sbjct 160 LQTDSVKGTFTIQDTGVGMNKEDLVSNLGTI 190
> pfa:PF11_0188 heat shock protein 90, putative
Length=930
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 37/90 (41%), Positives = 63/90 (70%), Gaps = 0/90 (0%)
Query 20 ELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRL 79
E KA+ ++L+ ++ ++ Y++KE+F+RELISN+SDA++K+R+ + K I++
Sbjct 74 EFKAETKKLLQIVAHSLYTDKEVFIRELISNSSDAIEKLRFLLQSGNIKASENITFHIKV 133
Query 80 IPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
D+ NN IEDSG+GM K E+++NLGT+
Sbjct 134 STDENNNLFIIEDSGVGMNKEEIIDNLGTI 163
> tgo:TGME49_092920 heat shock protein 90, putative ; K09488 TNF
receptor-associated protein 1
Length=861
Score = 78.6 bits (192), Expect = 5e-15, Method: Composition-based stats.
Identities = 44/105 (41%), Positives = 69/105 (65%), Gaps = 6/105 (5%)
Query 8 IDESEEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPE 67
+ +SE E F KA+ ++L+ ++ ++ Y++KE+F+RELISNA+DAL+K+R+ T
Sbjct 153 VADSEGEVHTF---KAETKKLLHIVTHSLYTDKEVFVRELISNAADALEKLRFLQATAQV 209
Query 68 KLKTKPE---LFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
E L I L D A T T++D+G+GMTKAEL+ +LGT+
Sbjct 210 TDADGSEAMALEIHLSTDAAAKTFTLQDTGVGMTKAELLEHLGTI 254
> pfa:PF14_0417 HSP90
Length=927
Score = 78.2 bits (191), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 57/143 (39%), Positives = 79/143 (55%), Gaps = 39/143 (27%)
Query 3 KEGLEIDESEEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIR--- 59
+E + D S EK F KA++ ++M +I+N+ Y++K++FLRELISNASDA DK R
Sbjct 89 REDISSDSSPVEKYNF---KAEVNKVMDIIVNSLYTDKDVFLRELISNASDACDKKRIIL 145
Query 60 --------YEAIT------EPEKLKTK-------------------PELFIRLIPDKANN 86
E +T E EK KT+ +L I++ PDK
Sbjct 146 ENNKLIKDAEVVTNEEIKNETEKEKTENVNESTDKKENVEEEKNDIKKLIIKIKPDKEKK 205
Query 87 TLTIEDSGIGMTKAELVNNLGTL 109
TLTI D+GIGM K+EL+NNLGT+
Sbjct 206 TLTITDNGIGMDKSELINNLGTI 228
> cel:R151.7 hypothetical protein
Length=479
Score = 72.4 bits (176), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 39/91 (42%), Positives = 60/91 (65%), Gaps = 3/91 (3%)
Query 19 EELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIR 78
E +A+ + LM ++ + YS+ E+F+RELISNASDAL+K RY + + + + E IR
Sbjct 45 HEFQAETRNLMDIVAKSLYSHSEVFVRELISNASDALEKRRYAEL-KGDVAEGPSE--IR 101
Query 79 LIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
+ +K T+T ED+GIGM + +LV LGT+
Sbjct 102 ITTNKDKRTITFEDTGIGMNREDLVKFLGTI 132
> hsa:29850 TRPM5, LTRPC5, MTR1; transient receptor potential
cation channel, subfamily M, member 5; K04980 transient receptor
potential cation channel subfamily M member 5
Length=1165
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 24/96 (25%), Positives = 46/96 (47%), Gaps = 6/96 (6%)
Query 6 LEIDESEEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITE 65
++I +SE E D++++M ++ SNK F+R + N +D D + Y + E
Sbjct 361 VDIAKSEIFNGDVEWKSCDLEEVM---VDALVSNKPEFVRLFVDNGADVADFLTYGRLQE 417
Query 66 PEKLKTKPELFIRLIPDKANNT-LTIEDSGIGMTKA 100
+ ++ L L+ K LT+ +G+G +A
Sbjct 418 LYRSVSRKSLLFDLLQRKQEEARLTL--AGLGTQQA 451
> dre:563561 myc box-dependent-interacting protein 1-like
Length=530
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 43/80 (53%), Gaps = 18/80 (22%)
Query 3 KEGLEIDESEEE----KKKFEELKADIQQLMSLIINT---FYSN--------KEIFLREL 47
KEG++I ++EEE +K FEE+ D+Q+ + + N+ FY N +E F RE+
Sbjct 167 KEGVKITKAEEELERAQKVFEEINIDLQEELPSLWNSRVGFYVNTFQSVAGLEEKFHREM 226
Query 48 I---SNASDALDKIRYEAIT 64
N D LDK+ + +T
Sbjct 227 GKINQNLYDTLDKLENQDMT 246
Lambda K H
0.312 0.133 0.354
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2067351240
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40