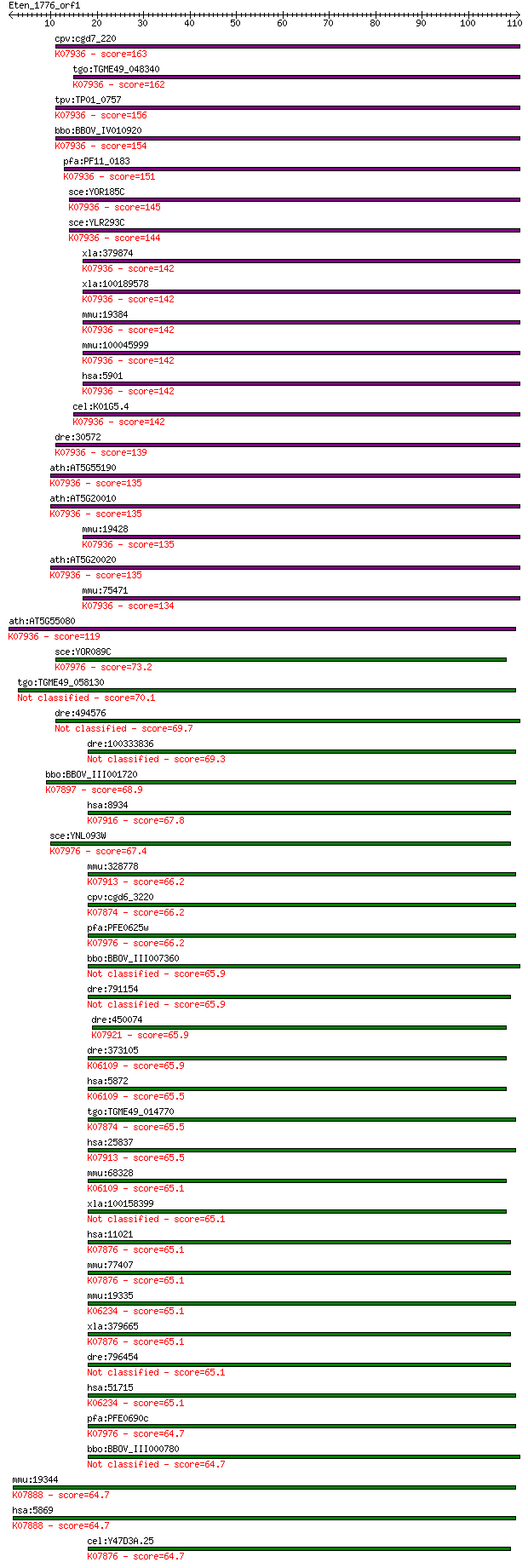
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1776_orf1
Length=110
Score E
Sequences producing significant alignments: (Bits) Value
cpv:cgd7_220 GTP-binding nuclear protein ran/tc4 ; K07936 GTP-... 163 1e-40
tgo:TGME49_048340 GTP-binding nuclear protein RAN/TC4, putativ... 162 3e-40
tpv:TP01_0757 GTP-binding nuclear protein ran; K07936 GTP-bind... 156 2e-38
bbo:BBOV_IV010920 23.m06098; GTP-binding nuclear protein Ran; ... 154 7e-38
pfa:PF11_0183 ran; GTP-binding nuclear protein ran/tc4; K07936... 151 5e-37
sce:YOR185C GSP2, CNR2; GTP binding protein (mammalian Ranp ho... 145 4e-35
sce:YLR293C GSP1, CNR1, CST17; Gsp1p; K07936 GTP-binding nucle... 144 6e-35
xla:379874 ran-1, MGC53579, ran; RAN, member RAS oncogene fami... 142 2e-34
xla:100189578 ran, ara24, gsp1, ran-1, tc4, xran; RAN, member ... 142 2e-34
mmu:19384 Ran; RAN, member RAS oncogene family; K07936 GTP-bin... 142 3e-34
mmu:100045999 GTP-binding nuclear protein Ran-like; K07936 GTP... 142 3e-34
hsa:5901 RAN, ARA24, Gsp1, TC4; RAN, member RAS oncogene famil... 142 3e-34
cel:K01G5.4 ran-1; associated with RAN (nuclear import/export)... 142 4e-34
dre:30572 ran, fc16b04, wu:fc16b04; ras-related nuclear protei... 139 2e-33
ath:AT5G55190 RAN3; RAN3 (RAN GTPASE 3); GTP binding / GTPase/... 135 3e-32
ath:AT5G20010 RAN-1; RAN-1; GTP binding / GTPase/ protein bind... 135 3e-32
mmu:19428 Rasl2-9-ps, M2, Ran, Ran/M2, Rasl2-9; RAS-like, fami... 135 3e-32
ath:AT5G20020 RAN2; RAN2; GTP binding / GTPase/ protein bindin... 135 4e-32
mmu:75471 1700009N14Rik, AI429145; RIKEN cDNA 1700009N14 gene;... 134 6e-32
ath:AT5G55080 AtRAN4; AtRAN4 (Ras-related nuclear protein 4); ... 119 2e-27
sce:YOR089C VPS21, VPS12, VPT12, YPT21, YPT51; GTPase required... 73.2 2e-13
tgo:TGME49_058130 Ras family domain-containing protein (EC:3.1... 70.1 2e-12
dre:494576 zgc:110195; si:dkey-13a21.4 69.7 2e-12
dre:100333836 RAB26, member RAS oncogene family-like 69.3 3e-12
bbo:BBOV_III001720 17.m07173; Rab7; K07897 Ras-related protein... 68.9 3e-12
hsa:8934 RAB7L1, DKFZp686P1051, RAB7L; RAB7, member RAS oncoge... 67.8 8e-12
sce:YNL093W YPT53; GTPase, similar to Ypt51p and Ypt52p and to... 67.4 1e-11
mmu:328778 Rab26, A830020M03Rik; RAB26, member RAS oncogene fa... 66.2 2e-11
cpv:cgd6_3220 RAS small GTpases RIC1/ypt1 ; K07874 Ras-related... 66.2 2e-11
pfa:PFE0625w Rab1b; Rab1b, GTPase; K07976 Rab family, other 66.2 2e-11
bbo:BBOV_III007360 17.m10679; Rab1a 65.9 3e-11
dre:791154 wu:fb77g03; zgc:158741 65.9 3e-11
dre:450074 rab34, zgc:101620; RAB34, member RAS oncogene famil... 65.9 3e-11
dre:373105 rab13, cb764, hm:zeh0455; RAB13, member RAS oncogen... 65.9 3e-11
hsa:5872 RAB13; RAB13, member RAS oncogene family; K06109 Ras-... 65.5 4e-11
tgo:TGME49_014770 RAS small GTpase, putative (EC:3.1.4.12); K0... 65.5 4e-11
hsa:25837 RAB26, V46133; RAB26, member RAS oncogene family; K0... 65.5 4e-11
mmu:68328 Rab13, 0610007N03Rik, B230212B15Rik; RAB13, member R... 65.1 5e-11
xla:100158399 hypothetical protein LOC100158399 65.1 5e-11
hsa:11021 RAB35, H-ray, RAB1C, RAY; RAB35, member RAS oncogene... 65.1 5e-11
mmu:77407 Rab35, 9530019H02Rik, AU040256, H-ray, RAB1C, RAY; R... 65.1 5e-11
mmu:19335 Rab23, AW545388, opb, opb2; RAB23, member RAS oncoge... 65.1 5e-11
xla:379665 rab35, MGC69101, h-ray, rab1c; RAB35, member RAS on... 65.1 5e-11
dre:796454 RAB35, member RAS oncogene family-like 65.1 6e-11
hsa:51715 RAB23, DKFZp781H0695, HSPC137, MGC8900; RAB23, membe... 65.1 6e-11
pfa:PFE0690c Rab1a; PfRab1a; K07976 Rab family, other 64.7 6e-11
bbo:BBOV_III000780 17.m07097; Rab1b 64.7 7e-11
mmu:19344 Rab5b, C030027M18Rik; RAB5B, member RAS oncogene fam... 64.7 7e-11
hsa:5869 RAB5B; RAB5B, member RAS oncogene family; K07888 Ras-... 64.7 7e-11
cel:Y47D3A.25 rab-35; RAB family member (rab-35); K07876 Ras-r... 64.7 7e-11
> cpv:cgd7_220 GTP-binding nuclear protein ran/tc4 ; K07936 GTP-binding
nuclear protein Ran
Length=212
Score = 163 bits (413), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 78/100 (78%), Positives = 85/100 (85%), Gaps = 7/100 (7%)
Query 11 MTEQVPEFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNF 70
M Q PEFKL LVGDGGVGKTTLVKRH G EFEKKYIPT+G EV PL+F+T+F
Sbjct 1 MAAQAPEFKLALVGDGGVGKTTLVKRHLTG-------EFEKKYIPTIGVEVHPLKFNTDF 53
Query 71 GPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
GP++FNVWDTAGQEKFGGLRDGYYIKGQCAI+MFDVTSRI
Sbjct 54 GPLIFNVWDTAGQEKFGGLRDGYYIKGQCAIIMFDVTSRI 93
> tgo:TGME49_048340 GTP-binding nuclear protein RAN/TC4, putative
(EC:3.1.4.12); K07936 GTP-binding nuclear protein Ran
Length=216
Score = 162 bits (409), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 80/96 (83%), Positives = 82/96 (85%), Gaps = 7/96 (7%)
Query 15 VPEFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIV 74
VPEFKLILVGDGGVGKTTLVKRH G EFEKKYIPTLG EV PL+F TNFG I
Sbjct 9 VPEFKLILVGDGGVGKTTLVKRHLTG-------EFEKKYIPTLGVEVHPLKFQTNFGMIT 61
Query 75 FNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
FNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVT+RI
Sbjct 62 FNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTARI 97
> tpv:TP01_0757 GTP-binding nuclear protein ran; K07936 GTP-binding
nuclear protein Ran
Length=211
Score = 156 bits (394), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 75/100 (75%), Positives = 83/100 (83%), Gaps = 7/100 (7%)
Query 11 MTEQVPEFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNF 70
M E+VP+FKL+LVGDGGVGKTTLVKRH G EFEKKYIPTLG EV PL+F TN
Sbjct 1 MAEEVPQFKLLLVGDGGVGKTTLVKRHLTG-------EFEKKYIPTLGVEVHPLKFRTNC 53
Query 71 GPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
G + FN WDTAGQEK+GGLRDGYYIKG+CAI+MFDVTSRI
Sbjct 54 GTVQFNAWDTAGQEKYGGLRDGYYIKGECAIIMFDVTSRI 93
> bbo:BBOV_IV010920 23.m06098; GTP-binding nuclear protein Ran;
K07936 GTP-binding nuclear protein Ran
Length=211
Score = 154 bits (389), Expect = 7e-38, Method: Compositional matrix adjust.
Identities = 75/100 (75%), Positives = 83/100 (83%), Gaps = 7/100 (7%)
Query 11 MTEQVPEFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNF 70
M E++P+FKL+LVGDGGVGKTTLVKRH G EFEKKYIPTLG EV PL+F TN
Sbjct 1 MAEEMPQFKLLLVGDGGVGKTTLVKRHLTG-------EFEKKYIPTLGVEVHPLKFRTNC 53
Query 71 GPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
G I FN WDTAGQEK+GGLRDGYYIKG+CAI+MFDVTSRI
Sbjct 54 GGIQFNAWDTAGQEKYGGLRDGYYIKGECAIIMFDVTSRI 93
> pfa:PF11_0183 ran; GTP-binding nuclear protein ran/tc4; K07936
GTP-binding nuclear protein Ran
Length=214
Score = 151 bits (381), Expect = 5e-37, Method: Compositional matrix adjust.
Identities = 73/98 (74%), Positives = 79/98 (80%), Gaps = 7/98 (7%)
Query 13 EQVPEFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGP 72
E +P++KLILVGDGGVGKTT VKRH G EFEKKYIPTLG EV PL+F TNFG
Sbjct 5 EYIPQYKLILVGDGGVGKTTFVKRHLTG-------EFEKKYIPTLGVEVHPLKFQTNFGK 57
Query 73 IVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
FNVWDTAGQEKFGGLRDGYYIK CAI+MFDV+SRI
Sbjct 58 TQFNVWDTAGQEKFGGLRDGYYIKSDCAIIMFDVSSRI 95
> sce:YOR185C GSP2, CNR2; GTP binding protein (mammalian Ranp
homolog) involved in the maintenance of nuclear organization,
RNA processing and transport; interacts with Kap121p, Kap123p
and Pdr6p (karyophilin betas); Gsp1p homolog that is not
required for viability; K07936 GTP-binding nuclear protein
Ran
Length=220
Score = 145 bits (365), Expect = 4e-35, Method: Compositional matrix adjust.
Identities = 71/97 (73%), Positives = 76/97 (78%), Gaps = 7/97 (7%)
Query 14 QVPEFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPI 73
+VP FKL+LVGDGG GKTT VKRH G EFEKKYI T+G EV PL F TNFG I
Sbjct 10 EVPTFKLVLVGDGGTGKTTFVKRHLTG-------EFEKKYIATIGVEVHPLSFYTNFGEI 62
Query 74 VFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
F+VWDTAGQEKFGGLRDGYYI QCAI+MFDVTSRI
Sbjct 63 KFDVWDTAGQEKFGGLRDGYYINAQCAIIMFDVTSRI 99
> sce:YLR293C GSP1, CNR1, CST17; Gsp1p; K07936 GTP-binding nuclear
protein Ran
Length=219
Score = 144 bits (364), Expect = 6e-35, Method: Compositional matrix adjust.
Identities = 71/97 (73%), Positives = 76/97 (78%), Gaps = 7/97 (7%)
Query 14 QVPEFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPI 73
+VP FKL+LVGDGG GKTT VKRH G EFEKKYI T+G EV PL F TNFG I
Sbjct 9 EVPTFKLVLVGDGGTGKTTFVKRHLTG-------EFEKKYIATIGVEVHPLSFYTNFGEI 61
Query 74 VFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
F+VWDTAGQEKFGGLRDGYYI QCAI+MFDVTSRI
Sbjct 62 KFDVWDTAGQEKFGGLRDGYYINAQCAIIMFDVTSRI 98
> xla:379874 ran-1, MGC53579, ran; RAN, member RAS oncogene family;
K07936 GTP-binding nuclear protein Ran
Length=216
Score = 142 bits (359), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 69/94 (73%), Positives = 75/94 (79%), Gaps = 7/94 (7%)
Query 17 EFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFN 76
+FKL+LVGDGG GKTT VKRH G EFEKKY+ TLG EV PL F TN GPI FN
Sbjct 10 QFKLVLVGDGGTGKTTFVKRHLTG-------EFEKKYVATLGVEVHPLVFHTNRGPIKFN 62
Query 77 VWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
VWDTAGQEKFGGLRDGYYI+ QCAI+MFDVTSR+
Sbjct 63 VWDTAGQEKFGGLRDGYYIQAQCAIIMFDVTSRV 96
> xla:100189578 ran, ara24, gsp1, ran-1, tc4, xran; RAN, member
RAS oncogene family; K07936 GTP-binding nuclear protein Ran
Length=216
Score = 142 bits (358), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 69/94 (73%), Positives = 75/94 (79%), Gaps = 7/94 (7%)
Query 17 EFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFN 76
+FKL+LVGDGG GKTT VKRH G EFEKKY+ TLG EV PL F TN GPI FN
Sbjct 10 QFKLVLVGDGGTGKTTFVKRHLTG-------EFEKKYVATLGVEVHPLVFHTNRGPIKFN 62
Query 77 VWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
VWDTAGQEKFGGLRDGYYI+ QCAI+MFDVTSR+
Sbjct 63 VWDTAGQEKFGGLRDGYYIQAQCAIIMFDVTSRV 96
> mmu:19384 Ran; RAN, member RAS oncogene family; K07936 GTP-binding
nuclear protein Ran
Length=216
Score = 142 bits (358), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 69/94 (73%), Positives = 75/94 (79%), Gaps = 7/94 (7%)
Query 17 EFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFN 76
+FKL+LVGDGG GKTT VKRH G EFEKKY+ TLG EV PL F TN GPI FN
Sbjct 10 QFKLVLVGDGGTGKTTFVKRHLTG-------EFEKKYVATLGVEVHPLVFHTNRGPIKFN 62
Query 77 VWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
VWDTAGQEKFGGLRDGYYI+ QCAI+MFDVTSR+
Sbjct 63 VWDTAGQEKFGGLRDGYYIQAQCAIIMFDVTSRV 96
> mmu:100045999 GTP-binding nuclear protein Ran-like; K07936 GTP-binding
nuclear protein Ran
Length=216
Score = 142 bits (358), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 69/94 (73%), Positives = 75/94 (79%), Gaps = 7/94 (7%)
Query 17 EFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFN 76
+FKL+LVGDGG GKTT VKRH G EFEKKY+ TLG EV PL F TN GPI FN
Sbjct 10 QFKLVLVGDGGTGKTTFVKRHLTG-------EFEKKYVATLGVEVHPLVFHTNRGPIKFN 62
Query 77 VWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
VWDTAGQEKFGGLRDGYYI+ QCAI+MFDVTSR+
Sbjct 63 VWDTAGQEKFGGLRDGYYIQAQCAIIMFDVTSRV 96
> hsa:5901 RAN, ARA24, Gsp1, TC4; RAN, member RAS oncogene family;
K07936 GTP-binding nuclear protein Ran
Length=216
Score = 142 bits (358), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 69/94 (73%), Positives = 75/94 (79%), Gaps = 7/94 (7%)
Query 17 EFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFN 76
+FKL+LVGDGG GKTT VKRH G EFEKKY+ TLG EV PL F TN GPI FN
Sbjct 10 QFKLVLVGDGGTGKTTFVKRHLTG-------EFEKKYVATLGVEVHPLVFHTNRGPIKFN 62
Query 77 VWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
VWDTAGQEKFGGLRDGYYI+ QCAI+MFDVTSR+
Sbjct 63 VWDTAGQEKFGGLRDGYYIQAQCAIIMFDVTSRV 96
> cel:K01G5.4 ran-1; associated with RAN (nuclear import/export)
function family member (ran-1); K07936 GTP-binding nuclear
protein Ran
Length=215
Score = 142 bits (357), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 69/96 (71%), Positives = 76/96 (79%), Gaps = 7/96 (7%)
Query 15 VPEFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIV 74
+P FKL+LVGDGG GKTT VKRH G EFEKKY+ TLG EV PL F TN G I
Sbjct 7 IPTFKLVLVGDGGTGKTTFVKRHLTG-------EFEKKYVATLGVEVHPLVFHTNRGQIR 59
Query 75 FNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
FNVWDTAGQEKFGGLRDGYYI+GQCAI+MFDVT+R+
Sbjct 60 FNVWDTAGQEKFGGLRDGYYIQGQCAIIMFDVTARV 95
> dre:30572 ran, fc16b04, wu:fc16b04; ras-related nuclear protein;
K07936 GTP-binding nuclear protein Ran
Length=215
Score = 139 bits (351), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 70/102 (68%), Positives = 77/102 (75%), Gaps = 9/102 (8%)
Query 11 MTEQVPE--FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFST 68
M E P+ FKL+LVGDGG GKTT VKRH G EFEKKY+ TLG EV PL F T
Sbjct 1 MAENEPQVQFKLVLVGDGGTGKTTFVKRHLTG-------EFEKKYVATLGVEVHPLVFHT 53
Query 69 NFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
N G I +NVWDTAGQEKFGGLRDGYYI+ QCAI+MFDVTSR+
Sbjct 54 NRGAIKYNVWDTAGQEKFGGLRDGYYIQAQCAIIMFDVTSRV 95
> ath:AT5G55190 RAN3; RAN3 (RAN GTPASE 3); GTP binding / GTPase/
protein binding; K07936 GTP-binding nuclear protein Ran
Length=221
Score = 135 bits (341), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 67/101 (66%), Positives = 74/101 (73%), Gaps = 7/101 (6%)
Query 10 KMTEQVPEFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTN 69
+ T P FKL++VGDGG GKTT VKRH G EFEKKY PT+G EV PL F TN
Sbjct 6 QQTVDYPSFKLVIVGDGGTGKTTFVKRHLTG-------EFEKKYEPTIGVEVHPLDFFTN 58
Query 70 FGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
G I F WDTAGQEKFGGLRDGYYI GQCAI+MFDVT+R+
Sbjct 59 CGKIRFYCWDTAGQEKFGGLRDGYYIHGQCAIIMFDVTARL 99
> ath:AT5G20010 RAN-1; RAN-1; GTP binding / GTPase/ protein binding;
K07936 GTP-binding nuclear protein Ran
Length=221
Score = 135 bits (340), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 67/101 (66%), Positives = 74/101 (73%), Gaps = 7/101 (6%)
Query 10 KMTEQVPEFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTN 69
+ T P FKL++VGDGG GKTT VKRH G EFEKKY PT+G EV PL F TN
Sbjct 6 QQTVDYPSFKLVIVGDGGTGKTTFVKRHLTG-------EFEKKYEPTIGVEVHPLDFFTN 58
Query 70 FGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
G I F WDTAGQEKFGGLRDGYYI GQCAI+MFDVT+R+
Sbjct 59 CGKIRFYCWDTAGQEKFGGLRDGYYIHGQCAIIMFDVTARL 99
> mmu:19428 Rasl2-9-ps, M2, Ran, Ran/M2, Rasl2-9; RAS-like, family
2, locus 9, pseudogene; K07936 GTP-binding nuclear protein
Ran
Length=216
Score = 135 bits (340), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 65/94 (69%), Positives = 74/94 (78%), Gaps = 7/94 (7%)
Query 17 EFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFN 76
+FK++LVGDGG GKTT +KRH G EFEK+Y+ TLG EV L F TN GPI FN
Sbjct 10 QFKVVLVGDGGTGKTTFMKRHLTG-------EFEKEYVATLGVEVHTLVFHTNRGPIKFN 62
Query 77 VWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
VWDTAGQEKFGGLRDGYYI+ QCAI+MFDVTSR+
Sbjct 63 VWDTAGQEKFGGLRDGYYIQAQCAIIMFDVTSRV 96
> ath:AT5G20020 RAN2; RAN2; GTP binding / GTPase/ protein binding;
K07936 GTP-binding nuclear protein Ran
Length=221
Score = 135 bits (340), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 67/101 (66%), Positives = 74/101 (73%), Gaps = 7/101 (6%)
Query 10 KMTEQVPEFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTN 69
+ T P FKL++VGDGG GKTT VKRH G EFEKKY PT+G EV PL F TN
Sbjct 6 QQTVDYPSFKLVIVGDGGTGKTTFVKRHLTG-------EFEKKYEPTIGVEVHPLDFFTN 58
Query 70 FGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
G I F WDTAGQEKFGGLRDGYYI GQCAI+MFDVT+R+
Sbjct 59 CGKIRFYCWDTAGQEKFGGLRDGYYIHGQCAIIMFDVTARL 99
> mmu:75471 1700009N14Rik, AI429145; RIKEN cDNA 1700009N14 gene;
K07936 GTP-binding nuclear protein Ran
Length=216
Score = 134 bits (337), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 67/94 (71%), Positives = 73/94 (77%), Gaps = 7/94 (7%)
Query 17 EFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFN 76
+FKL+LVGDGG GKT VKRH G EFEKKY+ TLG EV PL F T+ GPI FN
Sbjct 10 QFKLVLVGDGGTGKTAFVKRHLTG-------EFEKKYVATLGVEVHPLMFHTSRGPIKFN 62
Query 77 VWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
VWDTAGQEKFGGLRDGYYI+ Q AI+MFDVTSRI
Sbjct 63 VWDTAGQEKFGGLRDGYYIQAQGAIIMFDVTSRI 96
> ath:AT5G55080 AtRAN4; AtRAN4 (Ras-related nuclear protein 4);
GTP binding / GTPase/ protein binding; K07936 GTP-binding
nuclear protein Ran
Length=222
Score = 119 bits (298), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 61/109 (55%), Positives = 70/109 (64%), Gaps = 13/109 (11%)
Query 1 FPNPPPTDLKMTEQVPEFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAE 60
PN DL P FKL++VGDGG GKTT +KRH G EFE PTLG +
Sbjct 3 LPNQQNVDL------PTFKLLIVGDGGTGKTTFLKRHLTG-------EFEHNTEPTLGVD 49
Query 61 VRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSR 109
+ PL F TN G I F WDTAGQEK+ GL+D YYI GQCAI+MFDVT+R
Sbjct 50 IYPLDFFTNRGKIRFECWDTAGQEKYSGLKDAYYIHGQCAIIMFDVTAR 98
> sce:YOR089C VPS21, VPS12, VPT12, YPT21, YPT51; GTPase required
for transport during endocytosis and for correct sorting
of vacuolar hydrolases; localized in endocytic intermediates;
detected in mitochondria; geranylgeranylation required for
membrane association; mammalian Rab5 homolog; K07976 Rab family,
other
Length=210
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 36/97 (37%), Positives = 55/97 (56%), Gaps = 7/97 (7%)
Query 11 MTEQVPEFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNF 70
M V KL+L+G+ VGK+++V R F +++F + PT+GA R + N
Sbjct 1 MNTSVTSIKLVLLGEAAVGKSSIVLR-------FVSNDFAENKEPTIGAAFLTQRVTINE 53
Query 71 GPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVT 107
+ F +WDTAGQE+F L YY Q A++++DVT
Sbjct 54 HTVKFEIWDTAGQERFASLAPMYYRNAQAALVVYDVT 90
> tgo:TGME49_058130 Ras family domain-containing protein (EC:3.1.4.12)
Length=238
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 37/110 (33%), Positives = 59/110 (53%), Gaps = 10/110 (9%)
Query 3 NPPPTDLKMTEQVPE---FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGA 59
N P +++ ++P FKL+L+GD GVGK+ L+ R S A F + YI T+G
Sbjct 17 NWPLDRVQLCARIPLDHLFKLVLIGDSGVGKSCLLLRFSDDA-------FTESYITTIGV 69
Query 60 EVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSR 109
+ R + + + +WDTAGQE+F + YY ++++DVT R
Sbjct 70 DFRFRTINVDNEIVKLQIWDTAGQERFRTITSAYYRGADGIVLVYDVTDR 119
> dre:494576 zgc:110195; si:dkey-13a21.4
Length=205
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 35/101 (34%), Positives = 52/101 (51%), Gaps = 8/101 (7%)
Query 11 MTE-QVPEFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTN 69
MTE P KLIL+G+ GVGK++ + R F F Y T+G + + +
Sbjct 1 MTEWNTPHLKLILIGNSGVGKSSFMTR-------FVDHRFTNLYRATIGVDFLTKEITVD 53
Query 70 FGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
P++ +WDTAG E+F L Y C +++FDVTS +
Sbjct 54 RRPVILQIWDTAGTERFHSLGSSLYRGAHCCLLVFDVTSSV 94
> dre:100333836 RAB26, member RAS oncogene family-like
Length=148
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 35/92 (38%), Positives = 53/92 (57%), Gaps = 6/92 (6%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNV 77
FK++LVGD GVGKT L+ R GA L G+ +I T+G + R S + + +
Sbjct 56 FKVMLVGDSGVGKTCLLVRFKDGAFLAGS------FISTVGIDFRNKVLSIDSMRVKLQI 109
Query 78 WDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSR 109
WDTAGQE+F + YY ++++DVT++
Sbjct 110 WDTAGQERFRSVTHAYYRDAHALLLLYDVTNK 141
> bbo:BBOV_III001720 17.m07173; Rab7; K07897 Ras-related protein
Rab-7A
Length=233
Score = 68.9 bits (167), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 32/101 (31%), Positives = 54/101 (53%), Gaps = 7/101 (6%)
Query 9 LKMTEQVPEFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFST 68
+ M + P K+I++GD GVGKT+L+ N F F +Y T+GA+ +
Sbjct 17 ISMARKRPILKIIILGDSGVGKTSLM-------NQFINKRFTNQYRATIGADFSTQEVTV 69
Query 69 NFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSR 109
+ + +WDTAGQE+F L +Y C ++++D T++
Sbjct 70 DDKVVTLQIWDTAGQERFQSLGKAFYRGADCCMLVYDTTNQ 110
> hsa:8934 RAB7L1, DKFZp686P1051, RAB7L; RAB7, member RAS oncogene
family-like 1; K07916 Ras-related protein Rab-7L1
Length=203
Score = 67.8 bits (164), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 34/93 (36%), Positives = 56/93 (60%), Gaps = 10/93 (10%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAE--VRPLRFSTNFGPIVF 75
FK+++VGD VGKT+LV+R+S + F K Y T+G + ++ L++S ++ +
Sbjct 8 FKVLVVGDAAVGKTSLVQRYSQDS-------FSKHYKSTVGVDFALKVLQWS-DYEIVRL 59
Query 76 NVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTS 108
+WD AGQE+F + YY ++MFDVT+
Sbjct 60 QLWDIAGQERFTSMTRLYYRDASACVIMFDVTN 92
> sce:YNL093W YPT53; GTPase, similar to Ypt51p and Ypt52p and
to mammalian rab5; required for vacuolar protein sorting and
endocytosis; K07976 Rab family, other
Length=220
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 37/101 (36%), Positives = 58/101 (57%), Gaps = 9/101 (8%)
Query 10 KMTEQVPEF--KLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFS 67
K T +P K++L+G+ VGK+++V R F + +F++ PT+GA R +
Sbjct 3 KHTAAIPTLTIKVVLLGESAVGKSSIVLR-------FVSDDFKESKEPTIGAAFLTKRIT 55
Query 68 TNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTS 108
+ I F +WDTAGQE+F L YY Q A+++FDVT+
Sbjct 56 RDGKVIKFEIWDTAGQERFAPLAPMYYRNAQAALVVFDVTN 96
> mmu:328778 Rab26, A830020M03Rik; RAB26, member RAS oncogene
family; K07913 Ras-related protein Rab-26
Length=260
Score = 66.2 bits (160), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 33/92 (35%), Positives = 51/92 (55%), Gaps = 6/92 (6%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNV 77
FK++LVGD GVGKT L+ R GA L G +I T+G + R + + +
Sbjct 68 FKVMLVGDSGVGKTCLLVRFKDGAFLAGT------FISTVGIDFRNKVLDVDGMKVKLQI 121
Query 78 WDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSR 109
WDTAGQE+F + YY ++++D+T++
Sbjct 122 WDTAGQERFRSVTHAYYRDAHALLLLYDITNK 153
> cpv:cgd6_3220 RAS small GTpases RIC1/ypt1 ; K07874 Ras-related
protein Rab-1A
Length=203
Score = 66.2 bits (160), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 33/92 (35%), Positives = 48/92 (52%), Gaps = 7/92 (7%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNV 77
FKL+L+GD GVGK+ L+ R F + YI T+G + + S + +
Sbjct 9 FKLLLIGDSGVGKSCLLLR-------FADDTYTDSYISTIGVDFKIRTISLENKTVKLQI 61
Query 78 WDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSR 109
WDTAGQE+F + YY I+++DVT R
Sbjct 62 WDTAGQERFRTITSSYYRGAHGIIIVYDVTDR 93
> pfa:PFE0625w Rab1b; Rab1b, GTPase; K07976 Rab family, other
Length=200
Score = 66.2 bits (160), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 33/92 (35%), Positives = 48/92 (52%), Gaps = 7/92 (7%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNV 77
FK++L+GD GVGK+ L+ R F + YI T+G + + I +
Sbjct 9 FKILLIGDSGVGKSCLLLR-------FADDTYTDSYISTIGVDFKIKTIEIEDKIIKLQI 61
Query 78 WDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSR 109
WDTAGQE+F + YY Q I+++DVT R
Sbjct 62 WDTAGQERFRTITSSYYRGAQGIIIVYDVTDR 93
> bbo:BBOV_III007360 17.m10679; Rab1a
Length=213
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 48/93 (51%), Gaps = 7/93 (7%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNV 77
FKL+L+GD GVGK+ ++ R F F YI T+G + R + +
Sbjct 13 FKLVLIGDSGVGKSCVLLR-------FADDTFTDSYITTIGVDFRFRTIEVEGRRVKLQI 65
Query 78 WDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
WDTAGQE+F + YY I++FD+T ++
Sbjct 66 WDTAGQERFRTITSAYYRGADAIIIVFDITDKL 98
> dre:791154 wu:fb77g03; zgc:158741
Length=184
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 37/92 (40%), Positives = 51/92 (55%), Gaps = 9/92 (9%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNV 77
FKL+L+GD GVGKT L+ R S A F +I T+G + + N I +
Sbjct 9 FKLLLIGDSGVGKTCLLFRFSEDA-------FNTTFISTIGIDFKIRTIELNGKKIKLQI 61
Query 78 WDTAGQEKFGGLRDGYYIKGQCAIMM-FDVTS 108
WDTAGQE+F + YY +G IM+ +D+TS
Sbjct 62 WDTAGQERFRTITTAYY-RGAMGIMLVYDITS 92
> dre:450074 rab34, zgc:101620; RAB34, member RAS oncogene family;
K07921 Ras-related protein Rab-34
Length=260
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 35/89 (39%), Positives = 47/89 (52%), Gaps = 7/89 (7%)
Query 19 KLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNVW 78
K+I+VGD VGKT L+ R F F+K Y T+G + RF P +W
Sbjct 55 KVIVVGDLAVGKTCLINR-------FCKDVFDKNYKATIGVDFEMERFEVLGVPFSLQLW 107
Query 79 DTAGQEKFGGLRDGYYIKGQCAIMMFDVT 107
DTAGQE+F + YY Q I++FD+T
Sbjct 108 DTAGQERFKCIASTYYRGAQAVIIVFDLT 136
> dre:373105 rab13, cb764, hm:zeh0455; RAB13, member RAS oncogene
family; K06109 Ras-related protein Rab-13
Length=200
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 33/90 (36%), Positives = 46/90 (51%), Gaps = 7/90 (7%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNV 77
FKL+L+GD GVGKT L+ R F F YI T+G + + + V
Sbjct 9 FKLLLIGDSGVGKTCLIIR-------FAEDNFNSTYISTIGIDFKVKTIEVEGKKVKLQV 61
Query 78 WDTAGQEKFGGLRDGYYIKGQCAIMMFDVT 107
WDTAGQE+F + YY I+++D+T
Sbjct 62 WDTAGQERFKTITTAYYRGAMGIILVYDIT 91
> hsa:5872 RAB13; RAB13, member RAS oncogene family; K06109 Ras-related
protein Rab-13
Length=203
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 34/90 (37%), Positives = 46/90 (51%), Gaps = 7/90 (7%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNV 77
FKL+L+GD GVGKT L+ R F F YI T+G + + I V
Sbjct 9 FKLLLIGDSGVGKTCLIIR-------FAEDNFNNTYISTIGIDFKIRTVDIEGKKIKLQV 61
Query 78 WDTAGQEKFGGLRDGYYIKGQCAIMMFDVT 107
WDTAGQE+F + YY I+++D+T
Sbjct 62 WDTAGQERFKTITTAYYRGAMGIILVYDIT 91
> tgo:TGME49_014770 RAS small GTpase, putative (EC:3.1.4.12);
K07874 Ras-related protein Rab-1A
Length=202
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 32/92 (34%), Positives = 49/92 (53%), Gaps = 7/92 (7%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNV 77
FKL+L+GD GVGK+ L+ R F + + YI T+G + + + + +
Sbjct 9 FKLLLIGDSGVGKSCLLLR-------FADDTYTESYISTIGVDFKIRTIDLDGKTVKLQI 61
Query 78 WDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSR 109
WDTAGQE+F + YY I+++DVT R
Sbjct 62 WDTAGQERFRTITSSYYRGAHGIIIVYDVTDR 93
> hsa:25837 RAB26, V46133; RAB26, member RAS oncogene family;
K07913 Ras-related protein Rab-26
Length=256
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 34/92 (36%), Positives = 51/92 (55%), Gaps = 6/92 (6%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNV 77
FK++LVGD GVGKT L+ R GA L G +I T+G + R + + +
Sbjct 64 FKVMLVGDSGVGKTCLLVRFKDGAFLAGT------FISTVGIDFRNKVLDVDGVKVKLQM 117
Query 78 WDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSR 109
WDTAGQE+F + YY ++++DVT++
Sbjct 118 WDTAGQERFRSVTHAYYRDAHALLLLYDVTNK 149
> mmu:68328 Rab13, 0610007N03Rik, B230212B15Rik; RAB13, member
RAS oncogene family; K06109 Ras-related protein Rab-13
Length=202
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 34/90 (37%), Positives = 46/90 (51%), Gaps = 7/90 (7%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNV 77
FKL+L+GD GVGKT L+ R F F YI T+G + + I V
Sbjct 9 FKLLLIGDSGVGKTCLIIR-------FAEDNFNSTYISTIGIDFKIRTVDIEGKRIKLQV 61
Query 78 WDTAGQEKFGGLRDGYYIKGQCAIMMFDVT 107
WDTAGQE+F + YY I+++D+T
Sbjct 62 WDTAGQERFKTITTAYYRGAMGIILVYDIT 91
> xla:100158399 hypothetical protein LOC100158399
Length=201
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 34/90 (37%), Positives = 46/90 (51%), Gaps = 7/90 (7%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNV 77
FKL+L+GD GVGKT L+ R F F YI T+G + + I V
Sbjct 9 FKLLLIGDSGVGKTCLIVR-------FSEDSFNSTYISTIGIDFKIRTTEIEGKKIKLQV 61
Query 78 WDTAGQEKFGGLRDGYYIKGQCAIMMFDVT 107
WDTAGQE+F + YY I+++D+T
Sbjct 62 WDTAGQERFKTITTAYYRGAMGIILVYDIT 91
> hsa:11021 RAB35, H-ray, RAB1C, RAY; RAB35, member RAS oncogene
family; K07876 Ras-related protein Rab-35
Length=152
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 33/91 (36%), Positives = 50/91 (54%), Gaps = 7/91 (7%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNV 77
FKL+++GD GVGK++L+ R F + F YI T+G + + N + +
Sbjct 9 FKLLIIGDSGVGKSSLLLR-------FADNTFSGSYITTIGVDFKIRTVEINGEKVKLQI 61
Query 78 WDTAGQEKFGGLRDGYYIKGQCAIMMFDVTS 108
WDTAGQE+F + YY I+++DVTS
Sbjct 62 WDTAGQERFRTITSTYYRGTHGVIVVYDVTS 92
> mmu:77407 Rab35, 9530019H02Rik, AU040256, H-ray, RAB1C, RAY;
RAB35, member RAS oncogene family; K07876 Ras-related protein
Rab-35
Length=201
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 33/91 (36%), Positives = 50/91 (54%), Gaps = 7/91 (7%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNV 77
FKL+++GD GVGK++L+ R F + F YI T+G + + N + +
Sbjct 9 FKLLIIGDSGVGKSSLLLR-------FADNTFSGSYITTIGVDFKIRTVEINGEKVKLQI 61
Query 78 WDTAGQEKFGGLRDGYYIKGQCAIMMFDVTS 108
WDTAGQE+F + YY I+++DVTS
Sbjct 62 WDTAGQERFRTITSTYYRGTHGVIVVYDVTS 92
> mmu:19335 Rab23, AW545388, opb, opb2; RAB23, member RAS oncogene
family; K06234 Ras-related protein Rab-23
Length=237
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 29/92 (31%), Positives = 49/92 (53%), Gaps = 7/92 (7%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNV 77
K+++VG+G VGK+++++R+ G F K Y T+G + + N + +
Sbjct 10 IKMVVVGNGAVGKSSMIQRYCKGI-------FTKDYKKTIGVDFLERQIQVNDEDVRLML 62
Query 78 WDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSR 109
WDTAGQE+F + YY Q +++F T R
Sbjct 63 WDTAGQEEFDAITKAYYRGAQACVLVFSTTDR 94
> xla:379665 rab35, MGC69101, h-ray, rab1c; RAB35, member RAS
oncogene family; K07876 Ras-related protein Rab-35
Length=201
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 33/91 (36%), Positives = 50/91 (54%), Gaps = 7/91 (7%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNV 77
FKL+++GD GVGK++L+ R F + F YI T+G + + N + +
Sbjct 9 FKLLIIGDSGVGKSSLLLR-------FADNTFSGSYITTIGVDFKIRTVEINGEKVKLQI 61
Query 78 WDTAGQEKFGGLRDGYYIKGQCAIMMFDVTS 108
WDTAGQE+F + YY I+++DVTS
Sbjct 62 WDTAGQERFRTITSTYYRGTHGVIVVYDVTS 92
> dre:796454 RAB35, member RAS oncogene family-like
Length=199
Score = 65.1 bits (157), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 33/91 (36%), Positives = 50/91 (54%), Gaps = 7/91 (7%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNV 77
FKL+++GD GVGK++L+ R F + F YI T+G + + N + +
Sbjct 9 FKLLIIGDSGVGKSSLLLR-------FADNTFSGSYITTIGVDFKIRTVEINGEKVKLQI 61
Query 78 WDTAGQEKFGGLRDGYYIKGQCAIMMFDVTS 108
WDTAGQE+F + YY I+++DVTS
Sbjct 62 WDTAGQERFRTITSTYYRGTHGVIVVYDVTS 92
> hsa:51715 RAB23, DKFZp781H0695, HSPC137, MGC8900; RAB23, member
RAS oncogene family; K06234 Ras-related protein Rab-23
Length=237
Score = 65.1 bits (157), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 29/92 (31%), Positives = 49/92 (53%), Gaps = 7/92 (7%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNV 77
K+++VG+G VGK+++++R+ G F K Y T+G + + N + +
Sbjct 10 IKMVVVGNGAVGKSSMIQRYCKGI-------FTKDYKKTIGVDFLERQIQVNDEDVRLML 62
Query 78 WDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSR 109
WDTAGQE+F + YY Q +++F T R
Sbjct 63 WDTAGQEEFDAITKAYYRGAQACVLVFSTTDR 94
> pfa:PFE0690c Rab1a; PfRab1a; K07976 Rab family, other
Length=207
Score = 64.7 bits (156), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 48/92 (52%), Gaps = 7/92 (7%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNV 77
+K+IL+GD GVGK+ ++ R F F + YI T+G + R + + +
Sbjct 13 YKIILIGDSGVGKSCILLR-------FSDDHFTESYITTIGVDFRFRTIKVDDKIVKLQI 65
Query 78 WDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSR 109
WDTAGQE+F + YY I+++D T R
Sbjct 66 WDTAGQERFRTITSAYYRGADGIIIIYDTTDR 97
> bbo:BBOV_III000780 17.m07097; Rab1b
Length=221
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 31/93 (33%), Positives = 50/93 (53%), Gaps = 7/93 (7%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNV 77
FK+I++GD G GK++L+ R F + + Y+ T+G + + + I +
Sbjct 8 FKIIIIGDSGAGKSSLLLR-------FADDTYSESYMSTIGVDFKIKTVKIDNVTIKLQI 60
Query 78 WDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRI 110
WDTAGQE+F + YY I ++DVTSR+
Sbjct 61 WDTAGQERFRTITSTYYRGAHGIITVYDVTSRV 93
> mmu:19344 Rab5b, C030027M18Rik; RAB5B, member RAS oncogene family;
K07888 Ras-related protein Rab-5B
Length=215
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 36/108 (33%), Positives = 57/108 (52%), Gaps = 11/108 (10%)
Query 2 PNPPPTDLKMTEQVPEFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEV 61
PN P K + +FKL+L+G+ VGK++LV R G +F + T+GA
Sbjct 9 PNGQPQASK----ICQFKLVLLGESAVGKSSLVLRFVKG-------QFHEYQESTIGAAF 57
Query 62 RPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSR 109
+ + F +WDTAGQE++ L YY Q AI+++D+T++
Sbjct 58 LTQSVCLDDTTVKFEIWDTAGQERYHSLAPMYYRGAQAAIVVYDITNQ 105
> hsa:5869 RAB5B; RAB5B, member RAS oncogene family; K07888 Ras-related
protein Rab-5B
Length=215
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 36/108 (33%), Positives = 57/108 (52%), Gaps = 11/108 (10%)
Query 2 PNPPPTDLKMTEQVPEFKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEV 61
PN P K + +FKL+L+G+ VGK++LV R G +F + T+GA
Sbjct 9 PNGQPQASK----ICQFKLVLLGESAVGKSSLVLRFVKG-------QFHEYQESTIGAAF 57
Query 62 RPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSR 109
+ + F +WDTAGQE++ L YY Q AI+++D+T++
Sbjct 58 LTQSVCLDDTTVKFEIWDTAGQERYHSLAPMYYRGAQAAIVVYDITNQ 105
> cel:Y47D3A.25 rab-35; RAB family member (rab-35); K07876 Ras-related
protein Rab-35
Length=209
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 31/91 (34%), Positives = 51/91 (56%), Gaps = 7/91 (7%)
Query 18 FKLILVGDGGVGKTTLVKRHSHGANLFGASEFEKKYIPTLGAEVRPLRFSTNFGPIVFNV 77
FKL+++GD GVGK++L+ R F + F + YI T+G + + N + +
Sbjct 11 FKLLIIGDSGVGKSSLLLR-------FADNTFSENYITTIGVDFKIRTMDINGQRVKLQI 63
Query 78 WDTAGQEKFGGLRDGYYIKGQCAIMMFDVTS 108
WDTAGQE+F + YY ++++DVT+
Sbjct 64 WDTAGQERFRTITSTYYRGTHGVVVVYDVTN 94
Lambda K H
0.321 0.142 0.440
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2067351240
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40