bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1749_orf1
Length=111
Score E
Sequences producing significant alignments: (Bits) Value
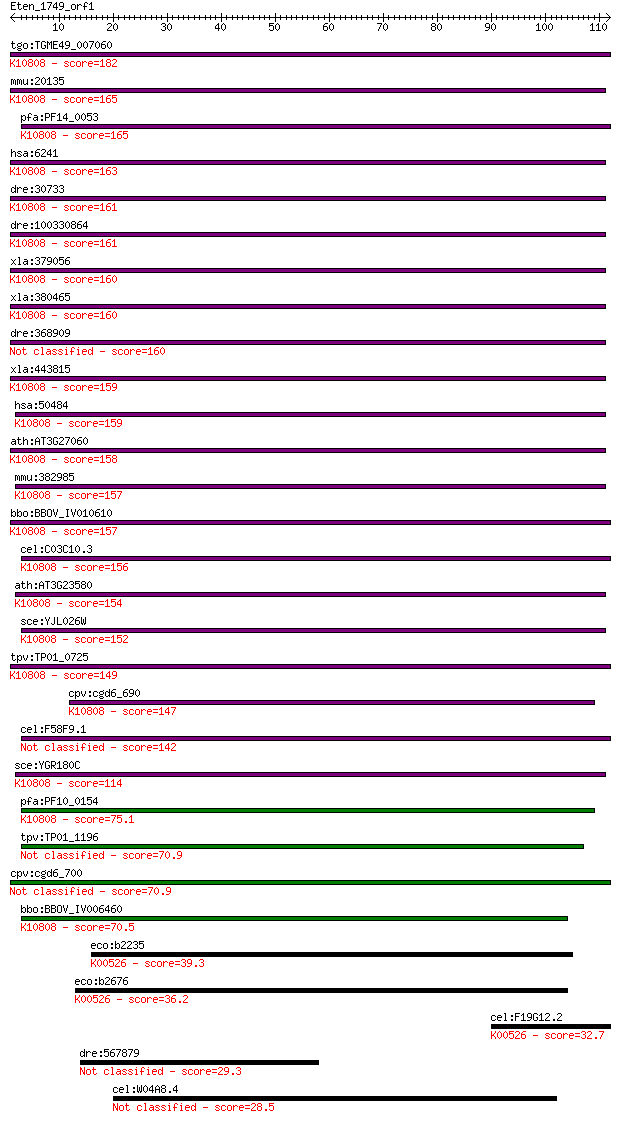
tgo:TGME49_007060 ribonucleotide-diphosphate reductase, small ... 182 3e-46
mmu:20135 Rrm2, AA407299, MGC113712, R2; ribonucleotide reduct... 165 4e-41
pfa:PF14_0053 ribonucleotide reductase small subunit; K10808 r... 165 4e-41
hsa:6241 RRM2, R2, RR2, RR2M; ribonucleotide reductase M2 (EC:... 163 2e-40
dre:30733 rrm2, cb111, chunp6884, r2; ribonucleotide reductase... 161 6e-40
dre:100330864 ribonucleoside-diphosphate reductase subunit M2-... 161 6e-40
xla:379056 rrm2b, MGC52676, p53r2; ribonucleotide reductase M2... 160 6e-40
xla:380465 rrm2.2, MGC53274, rrm2; ribonucleotide reductase M2... 160 7e-40
dre:368909 rrm2b, MGC123064, fb72h01, fb93b05, rrm2, si:zc215i... 160 1e-39
xla:443815 rrm2.1, MGC132324, MGC78958, rr2m; ribonucleotide r... 159 2e-39
hsa:50484 RRM2B, DKFZp686M05248, MGC102856, MGC42116, MTDPS8A,... 159 2e-39
ath:AT3G27060 TSO2; TSO2 (TSO meaning 'ugly' in Chinese); oxid... 158 5e-39
mmu:382985 Rrm2b, p53R2; ribonucleotide reductase M2 B (TP53 i... 157 8e-39
bbo:BBOV_IV010610 23.m06273; ribonucleoside-diphosphate reduct... 157 1e-38
cel:C03C10.3 rnr-2; RiboNucleotide Reductase family member (rn... 156 2e-38
ath:AT3G23580 RNR2A; RNR2A (RIBONUCLEOTIDE REDUCTASE 2A); ribo... 154 9e-38
sce:YJL026W RNR2, CRT6; Ribonucleotide-diphosphate reductase (... 152 3e-37
tpv:TP01_0725 ribonucleotide-diphosphate reductase small subun... 149 3e-36
cpv:cgd6_690 ribonucleotide reductase small subunit, duplicate... 147 9e-36
cel:F58F9.1 hypothetical protein 142 2e-34
sce:YGR180C RNR4, CRT3, PSO3; Ribonucleotide-diphosphate reduc... 114 9e-26
pfa:PF10_0154 ribonucleotide reductase small subunit, putative... 75.1 5e-14
tpv:TP01_1196 ribonucleotide-diphosphate reductase small subunit 70.9 9e-13
cpv:cgd6_700 ribonucleotide reductase small subunit 70.9 1e-12
bbo:BBOV_IV006460 23.m06516; ribonucleoside-diphosphate reduct... 70.5 1e-12
eco:b2235 nrdB, ECK2227, ftsB, JW2229; ribonucleoside-diphosph... 39.3 0.004
eco:b2676 nrdF, ECK2670, JW2651, ygaD; ribonucleoside-diphosph... 36.2 0.029
cel:F19G12.2 hypothetical protein; K00526 ribonucleoside-dipho... 32.7 0.29
dre:567879 vps13d; vacuolar protein sorting 13D (yeast) 29.3 2.9
cel:W04A8.4 hypothetical protein 28.5 6.1
> tgo:TGME49_007060 ribonucleotide-diphosphate reductase, small
subunit, putative (EC:1.17.4.1); K10808 ribonucleoside-diphosphate
reductase subunit M2 [EC:1.17.4.1]
Length=391
Score = 182 bits (461), Expect = 3e-46, Method: Compositional matrix adjust.
Identities = 80/111 (72%), Positives = 96/111 (86%), Gaps = 0/111 (0%)
Query 1 PRRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAA 60
P RWV+LPI HH +W MYKK EASFWT EEID++ D+ W TL NE+HFIK+VLAFFAA
Sbjct 80 PHRWVILPIQHHAIWEMYKKQEASFWTAEEIDLAQDMTHWETLGENEKHFIKYVLAFFAA 139
Query 61 SDGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRNE 111
SDGIV+ENLAE+F+ E+Q+PEAR+FY FQIA+ENIHSETYS+LID YIR+E
Sbjct 140 SDGIVLENLAEKFLTEIQVPEARAFYGFQIAMENIHSETYSLLIDQYIRDE 190
> mmu:20135 Rrm2, AA407299, MGC113712, R2; ribonucleotide reductase
M2 (EC:1.17.4.1); K10808 ribonucleoside-diphosphate reductase
subunit M2 [EC:1.17.4.1]
Length=390
Score = 165 bits (417), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 75/110 (68%), Positives = 89/110 (80%), Gaps = 0/110 (0%)
Query 1 PRRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAA 60
PRR+VV PI +H++W MYKKAEASFWT EE+D+S D+ W L +E+HFI HVLAFFAA
Sbjct 78 PRRFVVFPIEYHDIWQMYKKAEASFWTAEEVDLSKDIQHWEALKPDERHFISHVLAFFAA 137
Query 61 SDGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
SDGIV ENL ERF EVQ+ EAR FY FQIA+ENIHSE YS+LID YI++
Sbjct 138 SDGIVNENLVERFSQEVQVTEARCFYGFQIAMENIHSEMYSLLIDTYIKD 187
> pfa:PF14_0053 ribonucleotide reductase small subunit; K10808
ribonucleoside-diphosphate reductase subunit M2 [EC:1.17.4.1]
Length=349
Score = 165 bits (417), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 76/109 (69%), Positives = 95/109 (87%), Gaps = 0/109 (0%)
Query 3 RWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAASD 62
R+ + PI++ +VW+ YKKAEASFWT EEID+S+DL D+ L+ NE+HFIKHVLAFFAASD
Sbjct 40 RFTLYPILYPDVWDFYKKAEASFWTAEEIDLSSDLKDFEKLNENEKHFIKHVLAFFAASD 99
Query 63 GIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRNE 111
GIV+ENLA +F+ EVQI EA+ FY+FQIAVENIHSETYS+LID YI++E
Sbjct 100 GIVLENLASKFLREVQITEAKKFYSFQIAVENIHSETYSLLIDNYIKDE 148
> hsa:6241 RRM2, R2, RR2, RR2M; ribonucleotide reductase M2 (EC:1.17.4.1);
K10808 ribonucleoside-diphosphate reductase subunit
M2 [EC:1.17.4.1]
Length=389
Score = 163 bits (412), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 74/110 (67%), Positives = 89/110 (80%), Gaps = 0/110 (0%)
Query 1 PRRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAA 60
PRR+V+ PI +H++W MYKKAEASFWT EE+D+S D+ W +L E++FI HVLAFFAA
Sbjct 77 PRRFVIFPIEYHDIWQMYKKAEASFWTAEEVDLSKDIQHWESLKPEERYFISHVLAFFAA 136
Query 61 SDGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
SDGIV ENL ERF EVQI EAR FY FQIA+ENIHSE YS+LID YI++
Sbjct 137 SDGIVNENLVERFSQEVQITEARCFYGFQIAMENIHSEMYSLLIDTYIKD 186
> dre:30733 rrm2, cb111, chunp6884, r2; ribonucleotide reductase
M2 polypeptide (EC:1.17.4.1); K10808 ribonucleoside-diphosphate
reductase subunit M2 [EC:1.17.4.1]
Length=386
Score = 161 bits (407), Expect = 6e-40, Method: Compositional matrix adjust.
Identities = 73/110 (66%), Positives = 88/110 (80%), Gaps = 0/110 (0%)
Query 1 PRRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAA 60
P R+V+ PI +H++W MYKKAEASFWT EE+D+S DL W +L E++FI HVLAFFAA
Sbjct 74 PHRFVIFPIQYHDIWQMYKKAEASFWTAEEVDLSKDLQHWDSLKDEERYFISHVLAFFAA 133
Query 61 SDGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
SDGIV ENL ERF EVQ+ EAR FY FQIA+ENIHSE YS+LID YI++
Sbjct 134 SDGIVNENLVERFTQEVQVTEARCFYGFQIAMENIHSEMYSLLIDTYIKD 183
> dre:100330864 ribonucleoside-diphosphate reductase subunit M2-like;
K10808 ribonucleoside-diphosphate reductase subunit
M2 [EC:1.17.4.1]
Length=386
Score = 161 bits (407), Expect = 6e-40, Method: Compositional matrix adjust.
Identities = 73/110 (66%), Positives = 88/110 (80%), Gaps = 0/110 (0%)
Query 1 PRRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAA 60
P R+V+ PI +H++W MYKKAEASFWT EE+D+S DL W +L E++FI HVLAFFAA
Sbjct 74 PHRFVIFPIQYHDIWQMYKKAEASFWTAEEVDLSKDLQHWDSLKDEERYFISHVLAFFAA 133
Query 61 SDGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
SDGIV ENL ERF EVQ+ EAR FY FQIA+ENIHSE YS+LID YI++
Sbjct 134 SDGIVNENLVERFTQEVQVTEARCFYGFQIAMENIHSEMYSLLIDTYIKD 183
> xla:379056 rrm2b, MGC52676, p53r2; ribonucleotide reductase
M2 B (TP53 inducible) (EC:1.17.4.1); K10808 ribonucleoside-diphosphate
reductase subunit M2 [EC:1.17.4.1]
Length=386
Score = 160 bits (406), Expect = 6e-40, Method: Compositional matrix adjust.
Identities = 72/110 (65%), Positives = 88/110 (80%), Gaps = 0/110 (0%)
Query 1 PRRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAA 60
P R+V+ PI +H++W MYKKAEASFWT EE+D+S DL W +L E++FI HVLAFFAA
Sbjct 74 PHRFVIFPIQYHDIWQMYKKAEASFWTAEEVDLSKDLQHWESLKKEEKYFISHVLAFFAA 133
Query 61 SDGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
SDGIV ENL ERF EVQ+ EAR FY FQIA+ENIHSE YS+LID Y+++
Sbjct 134 SDGIVNENLVERFSKEVQVTEARCFYGFQIAMENIHSEMYSLLIDTYVKD 183
> xla:380465 rrm2.2, MGC53274, rrm2; ribonucleotide reductase
M2, gene 2 (EC:1.17.4.1); K10808 ribonucleoside-diphosphate
reductase subunit M2 [EC:1.17.4.1]
Length=386
Score = 160 bits (406), Expect = 7e-40, Method: Compositional matrix adjust.
Identities = 72/110 (65%), Positives = 89/110 (80%), Gaps = 0/110 (0%)
Query 1 PRRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAA 60
P R+V+ PI +H++W MYKKAEASFWT EE+D+S D+ W +L +E++FI HVLAFFAA
Sbjct 74 PHRFVIFPIQYHDIWQMYKKAEASFWTAEEVDLSKDIQHWESLKTDERYFISHVLAFFAA 133
Query 61 SDGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
SDGIV ENL ERF EVQ+ EAR FY FQIA+ENIHSE YS+LID YI++
Sbjct 134 SDGIVNENLVERFSKEVQVTEARCFYGFQIAMENIHSEMYSLLIDTYIKD 183
> dre:368909 rrm2b, MGC123064, fb72h01, fb93b05, rrm2, si:zc215i13.5,
wu:fb72h01, wu:fb93b05; ribonucleotide reductase M2
b (EC:1.17.4.1)
Length=349
Score = 160 bits (404), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 70/110 (63%), Positives = 89/110 (80%), Gaps = 0/110 (0%)
Query 1 PRRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAA 60
P+R+V+ PI + ++W MYK+A+ASFWT EE+D+S DL W L + E+HFI HVLAFFAA
Sbjct 37 PKRFVIFPIQYPDIWKMYKQAQASFWTVEEVDLSKDLTHWDGLKSEEKHFISHVLAFFAA 96
Query 61 SDGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
SDGIV ENL +RF EVQ+PEARSFY FQI +EN+HSE YS+LI+ YIR+
Sbjct 97 SDGIVNENLVQRFSQEVQLPEARSFYGFQILIENVHSEMYSMLINTYIRD 146
> xla:443815 rrm2.1, MGC132324, MGC78958, rr2m; ribonucleotide
reductase M2, gene 1 (EC:1.17.4.1); K10808 ribonucleoside-diphosphate
reductase subunit M2 [EC:1.17.4.1]
Length=406
Score = 159 bits (403), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 73/110 (66%), Positives = 88/110 (80%), Gaps = 0/110 (0%)
Query 1 PRRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAA 60
P R+V+LPI +H++W MYKKAEASFWT EE+D+S DL W TL E++FI +VLAFFAA
Sbjct 94 PGRFVILPIEYHDIWQMYKKAEASFWTAEEVDLSKDLQHWETLKPEERYFIAYVLAFFAA 153
Query 61 SDGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
SDGIV ENL ERF EVQ+ E R FY FQIA+ENIHSE YS+LID YI++
Sbjct 154 SDGIVNENLVERFSQEVQVTEVRCFYGFQIAMENIHSEMYSLLIDTYIKD 203
> hsa:50484 RRM2B, DKFZp686M05248, MGC102856, MGC42116, MTDPS8A,
MTDPS8B, P53R2; ribonucleotide reductase M2 B (TP53 inducible)
(EC:1.17.4.1); K10808 ribonucleoside-diphosphate reductase
subunit M2 [EC:1.17.4.1]
Length=423
Score = 159 bits (401), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 70/109 (64%), Positives = 88/109 (80%), Gaps = 0/109 (0%)
Query 2 RRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAAS 61
RR+V+ PI + ++W MYK+A+ASFWT EE+D+S DL W L A+E++FI H+LAFFAAS
Sbjct 112 RRFVIFPIQYPDIWKMYKQAQASFWTAEEVDLSKDLPHWNKLKADEKYFISHILAFFAAS 171
Query 62 DGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
DGIV ENL ERF EVQ+PEAR FY FQI +EN+HSE YS+LID YIR+
Sbjct 172 DGIVNENLVERFSQEVQVPEARCFYGFQILIENVHSEMYSLLIDTYIRD 220
> ath:AT3G27060 TSO2; TSO2 (TSO meaning 'ugly' in Chinese); oxidoreductase/
ribonucleoside-diphosphate reductase/ transition
metal ion binding (EC:1.17.4.1); K10808 ribonucleoside-diphosphate
reductase subunit M2 [EC:1.17.4.1]
Length=332
Score = 158 bits (399), Expect = 5e-39, Method: Compositional matrix adjust.
Identities = 72/111 (64%), Positives = 92/111 (82%), Gaps = 1/111 (0%)
Query 1 PRRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWA-TLDANEQHFIKHVLAFFA 59
P R+ + PI + ++W MYKKAEASFWT EE+D+S D DW +L+ E+HFIKHVLAFFA
Sbjct 14 PDRFCMFPIHYPQIWEMYKKAEASFWTAEEVDLSQDNRDWENSLNDGERHFIKHVLAFFA 73
Query 60 ASDGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
ASDGIV+ENLA RFM++VQ+ EAR+FY FQIA+ENIHSE YS+L+D YI++
Sbjct 74 ASDGIVLENLASRFMSDVQVSEARAFYGFQIAIENIHSEMYSLLLDTYIKD 124
> mmu:382985 Rrm2b, p53R2; ribonucleotide reductase M2 B (TP53
inducible) (EC:1.17.4.1); K10808 ribonucleoside-diphosphate
reductase subunit M2 [EC:1.17.4.1]
Length=351
Score = 157 bits (397), Expect = 8e-39, Method: Compositional matrix adjust.
Identities = 69/109 (63%), Positives = 88/109 (80%), Gaps = 0/109 (0%)
Query 2 RRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAAS 61
RR+V+ PI + ++W MYK+A+ASFWT EE+D+S DL W L ++E++FI H+LAFFAAS
Sbjct 40 RRFVIFPIQYPDIWRMYKQAQASFWTAEEVDLSKDLPHWNKLKSDEKYFISHILAFFAAS 99
Query 62 DGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
DGIV ENL ERF EVQ+PEAR FY FQI +EN+HSE YS+LID YIR+
Sbjct 100 DGIVNENLVERFSQEVQVPEARCFYGFQILIENVHSEMYSLLIDTYIRD 148
> bbo:BBOV_IV010610 23.m06273; ribonucleoside-diphosphate reductase
small subunit (EC:1.17.4.1); K10808 ribonucleoside-diphosphate
reductase subunit M2 [EC:1.17.4.1]
Length=339
Score = 157 bits (396), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 73/111 (65%), Positives = 90/111 (81%), Gaps = 0/111 (0%)
Query 1 PRRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAA 60
P R + PI + + W YKKA+ASFWT+EEID+S+DL DW TL E HFIK+VLAFFAA
Sbjct 28 PNRNGLYPIKYPDFWEWYKKAQASFWTSEEIDLSSDLKDWGTLTEGEHHFIKNVLAFFAA 87
Query 61 SDGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRNE 111
SDGIV+ENLA RF+ +V++PEA+ FY FQI VENIHSETYS+LI+ YIR+E
Sbjct 88 SDGIVLENLALRFLKDVKLPEAKFFYCFQITVENIHSETYSLLIEQYIRDE 138
> cel:C03C10.3 rnr-2; RiboNucleotide Reductase family member (rnr-2);
K10808 ribonucleoside-diphosphate reductase subunit
M2 [EC:1.17.4.1]
Length=381
Score = 156 bits (394), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 69/109 (63%), Positives = 86/109 (78%), Gaps = 0/109 (0%)
Query 3 RWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAASD 62
R+V+ P+ HH++WN YKKA ASFWT EE+D+ D+ DW ++ +EQ+FI +LAFFAASD
Sbjct 71 RFVIFPLKHHDIWNFYKKAVASFWTVEEVDLGKDMNDWEKMNGDEQYFISRILAFFAASD 130
Query 63 GIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRNE 111
GIV ENL ERF EVQ+ EAR FY FQIA+ENIHSE YS LI+ YIR+E
Sbjct 131 GIVNENLCERFSNEVQVSEARFFYGFQIAIENIHSEMYSKLIETYIRDE 179
> ath:AT3G23580 RNR2A; RNR2A (RIBONUCLEOTIDE REDUCTASE 2A); ribonucleoside-diphosphate
reductase (EC:1.17.4.1); K10808 ribonucleoside-diphosphate
reductase subunit M2 [EC:1.17.4.1]
Length=341
Score = 154 bits (388), Expect = 9e-38, Method: Compositional matrix adjust.
Identities = 66/109 (60%), Positives = 89/109 (81%), Gaps = 0/109 (0%)
Query 2 RRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAAS 61
+R+ + PI + +W MYKKAEASFWT EE+D+S D+ W L +E+HFI H+LAFFAAS
Sbjct 27 QRFTMFPIRYKSIWEMYKKAEASFWTAEEVDLSTDVQQWEALTDSEKHFISHILAFFAAS 86
Query 62 DGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
DGIV+ENLA RF+ +VQ+PEAR+FY FQIA+ENIHSE YS+L++ +I++
Sbjct 87 DGIVLENLAARFLNDVQVPEARAFYGFQIAMENIHSEMYSLLLETFIKD 135
> sce:YJL026W RNR2, CRT6; Ribonucleotide-diphosphate reductase
(RNR), small subunit; the RNR complex catalyzes the rate-limiting
step in dNTP synthesis and is regulated by DNA replication
and DNA damage checkpoint pathways via localization of
the small subunits (EC:1.17.4.1); K10808 ribonucleoside-diphosphate
reductase subunit M2 [EC:1.17.4.1]
Length=399
Score = 152 bits (383), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 72/109 (66%), Positives = 86/109 (78%), Gaps = 1/109 (0%)
Query 3 RWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWAT-LDANEQHFIKHVLAFFAAS 61
R V+ PI +HE+W YK+AEASFWT EEID+S D+ DW ++ NE+ FI VLAFFAAS
Sbjct 85 RTVLFPIKYHEIWQAYKRAEASFWTAEEIDLSKDIHDWNNRMNENERFFISRVLAFFAAS 144
Query 62 DGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
DGIV ENL E F EVQIPEA+SFY FQI +ENIHSETYS+LID YI++
Sbjct 145 DGIVNENLVENFSTEVQIPEAKSFYGFQIMIENIHSETYSLLIDTYIKD 193
> tpv:TP01_0725 ribonucleotide-diphosphate reductase small subunit;
K10808 ribonucleoside-diphosphate reductase subunit M2
[EC:1.17.4.1]
Length=308
Score = 149 bits (375), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 71/111 (63%), Positives = 90/111 (81%), Gaps = 0/111 (0%)
Query 1 PRRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAA 60
P R + PIV+ E W YKKA+ASFWT EEID S D W L+ +E+HFI +VLAFFAA
Sbjct 15 PNRTSLFPIVYPEFWEWYKKAQASFWTAEEIDFSMDYNHWHRLNKDERHFITNVLAFFAA 74
Query 61 SDGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRNE 111
SDGIV+ENLA +F+ +V+IPEA+SFY+FQIAVENIHSETYS+LI+ Y+++E
Sbjct 75 SDGIVLENLALKFLRDVKIPEAQSFYSFQIAVENIHSETYSLLIENYVKDE 125
> cpv:cgd6_690 ribonucleotide reductase small subunit, duplicated
adjacent gene ; K10808 ribonucleoside-diphosphate reductase
subunit M2 [EC:1.17.4.1]
Length=352
Score = 147 bits (370), Expect = 9e-36, Method: Compositional matrix adjust.
Identities = 68/97 (70%), Positives = 82/97 (84%), Gaps = 0/97 (0%)
Query 12 HEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAASDGIVMENLAE 71
++W+MYKKAEASFW TEEID+S D DW +L E+HFIK+VLAFFAASDGIVMENLA
Sbjct 52 KDIWSMYKKAEASFWVTEEIDLSQDTRDWESLKDPERHFIKYVLAFFAASDGIVMENLAV 111
Query 72 RFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYI 108
F+ E+QIPEAR +YAFQ+++E IHSETYS+LID YI
Sbjct 112 NFLREIQIPEARMYYAFQMSIEQIHSETYSLLIDRYI 148
> cel:F58F9.1 hypothetical protein
Length=581
Score = 142 bits (359), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 65/109 (59%), Positives = 82/109 (75%), Gaps = 0/109 (0%)
Query 3 RWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAASD 62
R+VV PIVH ++W YKKA A FWT+EE+D+ D+ W L ++E+ FI +LAFFAASD
Sbjct 316 RFVVHPIVHRDIWEFYKKAVACFWTSEEVDLGKDMSHWEILTSDERQFISSILAFFAASD 375
Query 63 GIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRNE 111
GIV ENL RF EVQ+ EAR FY FQIAVENIHSE Y+ L++ YIR++
Sbjct 376 GIVTENLCSRFSTEVQVTEARFFYGFQIAVENIHSEMYAKLLEAYIRDD 424
> sce:YGR180C RNR4, CRT3, PSO3; Ribonucleotide-diphosphate reductase
(RNR), small subunit; the RNR complex catalyzes the rate-limiting
step in dNTP synthesis and is regulated by DNA
replication and DNA damage checkpoint pathways via localization
of the small subunits (EC:1.17.4.1); K10808 ribonucleoside-diphosphate
reductase subunit M2 [EC:1.17.4.1]
Length=345
Score = 114 bits (284), Expect = 9e-26, Method: Compositional matrix adjust.
Identities = 50/109 (45%), Positives = 78/109 (71%), Gaps = 0/109 (0%)
Query 2 RRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAAS 61
RR+V+ PI +HE+W YKK EASFWT EEI+++ D D+ L +++ +I ++LA +S
Sbjct 33 RRFVMFPIKYHEIWAAYKKVEASFWTAEEIELAKDTEDFQKLTDDQKTYIGNLLALSISS 92
Query 62 DGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
D +V + L E F A++Q PE +SFY FQI +ENI+SE YS+++D + ++
Sbjct 93 DNLVNKYLIENFSAQLQNPEGKSFYGFQIMMENIYSEVYSMMVDAFFKD 141
> pfa:PF10_0154 ribonucleotide reductase small subunit, putative;
K10808 ribonucleoside-diphosphate reductase subunit M2 [EC:1.17.4.1]
Length=335
Score = 75.1 bits (183), Expect = 5e-14, Method: Composition-based stats.
Identities = 38/109 (34%), Positives = 60/109 (55%), Gaps = 3/109 (2%)
Query 3 RWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAASD 62
RWV+ PI + W+ YK+ E+ FWT E+ + D +D N + ++ F++ D
Sbjct 28 RWVMFPIKYKTFWSYYKEIESLFWTAEDYNFDKDKQYLENIDKNMLVKLFELICFYSLKD 87
Query 63 GIVMENLA---ERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYI 108
V E A + + +QIPE R+FY FQ+ +ENIH E Y+ + + YI
Sbjct 88 LHVYEEQALITSKMLDIIQIPEGRAFYGFQMCMENIHDEVYACIFETYI 136
> tpv:TP01_1196 ribonucleotide-diphosphate reductase small subunit
Length=326
Score = 70.9 bits (172), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 40/107 (37%), Positives = 56/107 (52%), Gaps = 3/107 (2%)
Query 3 RWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAASD 62
RWV+ PI H W MYK+ E +FW E+ S DL L + + H+L+F D
Sbjct 31 RWVMFPIEHDTFWIMYKEVENNFWAAEDFIFSDDLNSLFKLPKPLFNSLFHLLSFHINLD 90
Query 63 G---IVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDL 106
++ ++EVQI EAR+FY FQ+ ENIH+ET + L
Sbjct 91 NKWVCRPVDMTLELLSEVQIAEARAFYGFQLTDENIHTETIGTMFQL 137
> cpv:cgd6_700 ribonucleotide reductase small subunit
Length=339
Score = 70.9 bits (172), Expect = 1e-12, Method: Composition-based stats.
Identities = 35/113 (30%), Positives = 60/113 (53%), Gaps = 2/113 (1%)
Query 1 PRRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAA 60
P RWV+ PI + +W++YKK E SFWT E I D +D +I+ A
Sbjct 22 PFRWVLFPIKYSRLWSLYKKYEVSFWTAEVISPMEDKTKLMNMDEKLLDYIEMAFAKRIY 81
Query 61 SDGIVMEN--LAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRNE 111
D ++ L + +VQ+PEAR +++F + +ENI+ E + +++ +N+
Sbjct 82 IDNTCLDTVLLTCELLGQVQVPEARGYFSFTMCMENIYKELFMNYVEMIRKNK 134
> bbo:BBOV_IV006460 23.m06516; ribonucleoside-diphosphate reductase
beta subunit (EC:1.17.4.1); K10808 ribonucleoside-diphosphate
reductase subunit M2 [EC:1.17.4.1]
Length=329
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 34/104 (32%), Positives = 59/104 (56%), Gaps = 3/104 (2%)
Query 3 RWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAASD 62
RWV+ PI + +W MYK+ E SFW E+ + + +++L + + ++++ D
Sbjct 30 RWVMFPIHYDALWAMYKEIENSFWAAEDFRFANERDTYSSLPTELRQLVVKLISYHNRLD 89
Query 63 GIVMENLAE---RFMAEVQIPEARSFYAFQIAVENIHSETYSVL 103
+ A +A+ QIPEAR+FY FQ++ ENIHSE + ++
Sbjct 90 RSEVARPATITLDLLADTQIPEARAFYGFQVSYENIHSELFGIM 133
> eco:b2235 nrdB, ECK2227, ftsB, JW2229; ribonucleoside-diphosphate
reductase 1, beta subunit, ferritin-like protein (EC:1.17.4.1);
K00526 ribonucleoside-diphosphate reductase beta chain
[EC:1.17.4.1]
Length=376
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 24/89 (26%), Positives = 42/89 (47%), Gaps = 0/89 (0%)
Query 16 NMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAASDGIVMENLAERFMA 75
+ +K + FW EE+D+S D +D+ L +E+H L + D I + +
Sbjct 39 KLIEKQLSFFWRPEEVDVSRDRIDYQALPEHEKHIFISNLKYQTLLDSIQGRSPNVALLP 98
Query 76 EVQIPEARSFYAFQIAVENIHSETYSVLI 104
+ IPE ++ E IHS +Y+ +I
Sbjct 99 LISIPELETWVETWAFSETIHSRSYTHII 127
> eco:b2676 nrdF, ECK2670, JW2651, ygaD; ribonucleoside-diphosphate
reductase 2, beta subunit, ferritin-like protein (EC:1.17.4.1);
K00526 ribonucleoside-diphosphate reductase beta chain
[EC:1.17.4.1]
Length=319
Score = 36.2 bits (82), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 23/91 (25%), Positives = 39/91 (42%), Gaps = 3/91 (3%)
Query 13 EVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAASDGIVMENLAER 72
EVWN + ++FW E++ +S D+ W TL EQ V D + A
Sbjct 21 EVWN---RLTSNFWLPEKVPLSNDIPAWQTLTVVEQQLTMRVFTGLTLLDTLQNVIGAPS 77
Query 73 FMAEVQIPEARSFYAFQIAVENIHSETYSVL 103
M + P + + +E +H+ +YS +
Sbjct 78 LMPDALTPHEEAVLSNISFMEAVHARSYSSI 108
> cel:F19G12.2 hypothetical protein; K00526 ribonucleoside-diphosphate
reductase beta chain [EC:1.17.4.1]
Length=668
Score = 32.7 bits (73), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 14/22 (63%), Positives = 19/22 (86%), Gaps = 0/22 (0%)
Query 90 IAVENIHSETYSVLIDLYIRNE 111
IAVENIHSE Y+ L++ YIR++
Sbjct 445 IAVENIHSEMYAKLLEAYIRDD 466
> dre:567879 vps13d; vacuolar protein sorting 13D (yeast)
Length=4386
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 21/44 (47%), Gaps = 0/44 (0%)
Query 14 VWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAF 57
V+N+YK E F+ E DM L + + Q F V+AF
Sbjct 1978 VYNIYKYGETDFFLQREYDMKVSLQMASVQYVHTQRFQAEVVAF 2021
> cel:W04A8.4 hypothetical protein
Length=129
Score = 28.5 bits (62), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 22/82 (26%), Positives = 35/82 (42%), Gaps = 9/82 (10%)
Query 20 KAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAASDGIVMENLAERFMAEVQI 79
KAEA++WT ++ + DW H L +A D IVM + ++ + +
Sbjct 56 KAEATYWTGLDLHKGETMSDWTN-----THLRGDQLR-YAIMDTIVMHLIIKQRLRNIHK 109
Query 80 PEARSFYAFQIAVENIHSETYS 101
+ R FQ EN H Y+
Sbjct 110 NDPR---LFQYVFENGHPLPYN 128
Lambda K H
0.323 0.133 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2062416360
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40