bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1742_orf2
Length=177
Score E
Sequences producing significant alignments: (Bits) Value
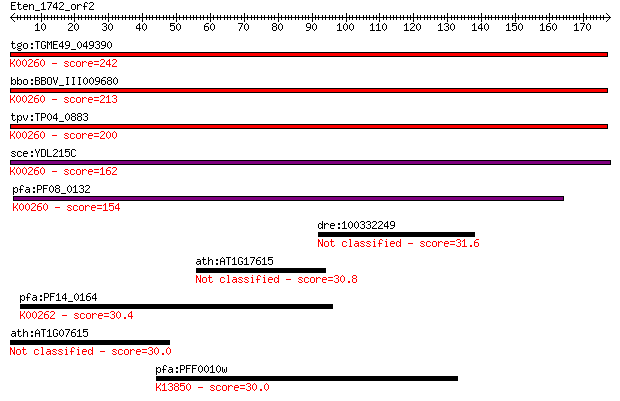
tgo:TGME49_049390 NAD-specific glutamate dehydrogenase, putati... 242 5e-64
bbo:BBOV_III009680 17.m07840; Glutamate/Leucine/Phenylalanine/... 213 2e-55
tpv:TP04_0883 NAD-specific glutamate dehydrogenase (EC:1.4.1.2... 200 2e-51
sce:YDL215C GDH2; Gdh2p (EC:1.4.1.2); K00260 glutamate dehydro... 162 6e-40
pfa:PF08_0132 glutamate dehydrogenase, putative (EC:1.4.1.2); ... 154 1e-37
dre:100332249 Extracellular matrix protein 1-like 31.6 1.6
ath:AT1G17615 disease resistance protein (TIR-NBS class), puta... 30.8 2.5
pfa:PF14_0164 NADP-specific glutamate dehydrogenase; K00262 gl... 30.4 3.2
ath:AT1G07615 GTP binding 30.0 4.1
pfa:PFF0010w VAR; erythrocyte membrane protein 1, PfEMP1; K138... 30.0 4.7
> tgo:TGME49_049390 NAD-specific glutamate dehydrogenase, putative
(EC:1.4.1.2); K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1113
Score = 242 bits (618), Expect = 5e-64, Method: Compositional matrix adjust.
Identities = 113/176 (64%), Positives = 139/176 (78%), Gaps = 0/176 (0%)
Query 1 AFTTGKRPENGGIPHDVFGMTTASVETYVRGIYEKLGLDEREMTRIQTGGPDGDLGANAL 60
AFTTGK P GGIPHD +GMTTAS+ETY+ GI EK L E E+TR GGPDGDLG+NAL
Sbjct 719 AFTTGKLPAMGGIPHDTYGMTTASIETYIHGILEKKNLKEEEVTRQLVGGPDGDLGSNAL 778
Query 61 LQTKSKTLAVVDGAGVVYDPEGLNAEELRRLARLRFKGEKTSSMLFDSSLLSPKGFKVPQ 120
L++ +KT ++VDG+GV++DPEGL+ ELRRLA+ RF+G +TS+ML+D LLSP GFKV Q
Sbjct 779 LKSNTKTTSIVDGSGVLHDPEGLDINELRRLAKRRFEGLQTSAMLYDEKLLSPMGFKVSQ 838
Query 121 DARDITLPDGTYVASGIEFRNNFHLSKFAVCDLFNPCGGRPASVNPRNVEQLFEQQ 176
D RD+ LPDGT VASG EFR FHL DLFNPCGGRPASV P NV+++F+++
Sbjct 839 DDRDVVLPDGTAVASGFEFRGRFHLDPRGSADLFNPCGGRPASVTPFNVDKMFDEK 894
> bbo:BBOV_III009680 17.m07840; Glutamate/Leucine/Phenylalanine/Valine
dehydrogenase family protein (EC:1.4.1.2); K00260 glutamate
dehydrogenase [EC:1.4.1.2]
Length=1025
Score = 213 bits (543), Expect = 2e-55, Method: Compositional matrix adjust.
Identities = 102/176 (57%), Positives = 132/176 (75%), Gaps = 1/176 (0%)
Query 1 AFTTGKRPENGGIPHDVFGMTTASVETYVRGIYEKLGLDEREMTRIQTGGPDGDLGANAL 60
+FTTGK P GGIPHD +GMTTAS+E Y+ + L+E E+TR TGGPDGDLG+NAL
Sbjct 646 SFTTGKEPVMGGIPHDTYGMTTASIEAYIHELLNIFHLNEEEVTRFLTGGPDGDLGSNAL 705
Query 61 LQTKSKTLAVVDGAGVVYDPEGLNAEELRRLARLRFKGEKTSSMLFDSSLLSPKGFKVPQ 120
L +K+KTL V+D +GV++DPEGL+ EL+RLA R KG TS+M ++ +LLS KGFKVP+
Sbjct 706 LCSKTKTLTVIDKSGVLHDPEGLDINELQRLAANRLKGLPTSAMHYNEALLSDKGFKVPE 765
Query 121 DARDITLPDGTYVASGIEFRNNFHLSKFAVCDLFNPCGGRPASVNPRNVEQLFEQQ 176
DA D+ LPDGT V G +FR+ FHL DLFNPCGGRP+S+ P NV +LF+++
Sbjct 766 DAVDMVLPDGTKVKRGHKFRDEFHLGA-CPSDLFNPCGGRPSSITPFNVNRLFDEK 820
> tpv:TP04_0883 NAD-specific glutamate dehydrogenase (EC:1.4.1.2);
K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1178
Score = 200 bits (508), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 102/212 (48%), Positives = 134/212 (63%), Gaps = 36/212 (16%)
Query 1 AFTTGKRPENGGIPHDVFGMTTASVETYVRGIYEKLGLDEREMTRIQTGGPDGDLGANAL 60
+FTTGK P+ GGIPHD++GMTT S+E YV GI K GL E E+TR TGGPDGDLG+NA+
Sbjct 734 SFTTGKAPQLGGIPHDIYGMTTTSIEAYVTGILNKYGLKEEEVTRFLTGGPDGDLGSNAI 793
Query 61 LQTKSKTLAVVDGAGVVYDPEGLNAEELRRLARLR------------------------- 95
+ +KTL V+D +GV++DP GL+ ELRRLA LR
Sbjct 794 KVSNTKTLTVLDKSGVLHDPNGLDLNELRRLAFLRDTTHLSATDNPNNVTLNHSNSSLQR 853
Query 96 ---FKGE--------KTSSMLFDSSLLSPKGFKVPQDARDITLPDGTYVASGIEFRNNFH 144
+G+ KT SM +D LLS KGF VP++A ++ LPDG V +G +FR+ FH
Sbjct 854 SSSLEGDEELLGARLKTCSMGYDKRLLSSKGFMVPEEAMNVVLPDGFVVKNGYKFRDEFH 913
Query 145 LSKFAVCDLFNPCGGRPASVNPRNVEQLFEQQ 176
LS +A DLF PCGGRP+S+ P NV +LF+++
Sbjct 914 LSSYAKADLFCPCGGRPSSITPFNVNRLFDEK 945
> sce:YDL215C GDH2; Gdh2p (EC:1.4.1.2); K00260 glutamate dehydrogenase
[EC:1.4.1.2]
Length=1092
Score = 162 bits (410), Expect = 6e-40, Method: Composition-based stats.
Identities = 86/182 (47%), Positives = 111/182 (60%), Gaps = 10/182 (5%)
Query 1 AFTTGKRPENGGIPHDVFGMTTASVETYVRGIYEKLGLDEREMTRIQTGGPDGDLGANAL 60
+F TGK P GGIPHD +GMT+ V YV IYE L L + + QTGGPDGDLG+N +
Sbjct 704 SFLTGKSPSLGGIPHDEYGMTSLGVRAYVNKIYETLNLTNSTVYKFQTGGPDGDLGSNEI 763
Query 61 LQTKSKT--LAVVDGAGVVYDPEGLNAEELRRLARLRFKGEKTSSMLFDSSLLSPKGFKV 118
L + LA++DG+GV+ DP+GL+ +EL RLA E+ FD+S LS GF V
Sbjct 764 LLSSPNECYLAILDGSGVLCDPKGLDKDELCRLAH-----ERKMISDFDTSKLSNNGFFV 818
Query 119 PQDARDITLPDGTYVASGIEFRNNFHLSKFAV---CDLFNPCGGRPASVNPRNVEQLFEQ 175
DA DI LP+GT VA+G FRN FH F D+F PCGGRP S+ N+ ++
Sbjct 819 SVDAMDIMLPNGTIVANGTTFRNTFHTQIFKFVDHVDIFVPCGGRPNSITLNNLHYFVDE 878
Query 176 QT 177
+T
Sbjct 879 KT 880
> pfa:PF08_0132 glutamate dehydrogenase, putative (EC:1.4.1.2);
K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1397
Score = 154 bits (390), Expect = 1e-37, Method: Composition-based stats.
Identities = 76/166 (45%), Positives = 116/166 (69%), Gaps = 5/166 (3%)
Query 2 FTTGKRPENGGIPHDVFGMTTASVETYVRGIYEKLGLDEREMTRIQTGGPDGDLGANALL 61
F+TGK +NGG+PHD++GMTT +ETY+ + EKL + E ++R GGPDGDLG+NA+L
Sbjct 936 FSTGKLRKNGGVPHDMYGMTTLGIETYISKLCEKLNIKEESISRSLVGGPDGDLGSNAIL 995
Query 62 QTKSKTLAVVDGAGVVYDPEGLNAEELRRLARLRFKGEKTSSM----LFDSSLLSPKGFK 117
Q+K+K ++++DG+G++YD +GLN EEL RLA+ R +K+ ++ L+D S GFK
Sbjct 996 QSKTKIISIIDGSGILYDKQGLNKEELIRLAKRRNNKDKSKAITCCTLYDEKYFSKDGFK 1055
Query 118 VPQDARDITLPDGTYVASGIEFRNNFHLSKFAVCDLFNPCGGRPAS 163
+ + ++ + G + +G++FRN F L+ C+LFNPCGGRP S
Sbjct 1056 ISIEDHNVDI-FGNKIRNGLDFRNTFFLNPLNKCELFNPCGGRPHS 1100
> dre:100332249 Extracellular matrix protein 1-like
Length=499
Score = 31.6 bits (70), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 23/46 (50%), Gaps = 0/46 (0%)
Query 92 ARLRFKGEKTSSMLFDSSLLSPKGFKVPQDARDITLPDGTYVASGI 137
A +FKG K +S SL SPK K A D+T P G +S I
Sbjct 238 ASQKFKGFKYNSSACKGSLASPKALKKQTAAPDLTFPPGRPESSNI 283
> ath:AT1G17615 disease resistance protein (TIR-NBS class), putative
Length=380
Score = 30.8 bits (68), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 56 GANALLQTKSKTLAVVDGAGVVYDPEGLNAEELRRLAR 93
G+ L+ T+ K L V G VVY+ E L E+R+L R
Sbjct 318 GSIVLITTQDKQLLVAFGIKVVYEVECLRCFEVRQLFR 355
> pfa:PF14_0164 NADP-specific glutamate dehydrogenase; K00262
glutamate dehydrogenase (NADP+) [EC:1.4.1.4]
Length=470
Score = 30.4 bits (67), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 26/93 (27%), Positives = 39/93 (41%), Gaps = 2/93 (2%)
Query 4 TGKRPENGGIPHDVFGMTTASVETYVRGIYEKLGLDEREMTRIQTGGPDGDL-GANALLQ 62
TGK + GG V T + +V + + L + + T + +G + L LL
Sbjct 215 TGKNVKWGGSNLRV-EATGYGLVYFVLEVLKSLNIPVEKQTAVVSGSGNVALYCVQKLLH 273
Query 63 TKSKTLAVVDGAGVVYDPEGLNAEELRRLARLR 95
K L + D G VY+P G E L L L+
Sbjct 274 LNVKVLTLSDSNGYVYEPNGFTHENLEFLIDLK 306
> ath:AT1G07615 GTP binding
Length=493
Score = 30.0 bits (66), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 25/47 (53%), Gaps = 1/47 (2%)
Query 1 AFTTGKRPENGGIPHDVFGMTTASVETYVRGIYEKLGLDEREMTRIQ 47
AFTT RP G + +D F MT A + ++G ++ GL + I+
Sbjct 334 AFTT-LRPNLGNVNYDDFSMTVADIPGLIKGAHQNRGLGHNFLRHIE 379
> pfa:PFF0010w VAR; erythrocyte membrane protein 1, PfEMP1; K13850
erythrocyte membrane protein 1
Length=2879
Score = 30.0 bits (66), Expect = 4.7, Method: Composition-based stats.
Identities = 33/99 (33%), Positives = 44/99 (44%), Gaps = 12/99 (12%)
Query 44 TRIQTGGPDG---DLGANALLQTKSKTLAVVDGAGVVYDPEG-LNAEELRRLARL----- 94
T +QT P G LG+ A L K KTL+ VD V+ P+G N L+ R
Sbjct 2455 TILQTTIPFGVALALGSIAFLFLKKKTLSPVDLFSVINIPKGDYNIPTLKSSNRYIPYAS 2514
Query 95 -RFKGEKTSSMLFDSSLLSPKGFKVPQDARDITLPDGTY 132
R+KG+ M DSS + + D DIT + Y
Sbjct 2515 DRYKGKTYIYMEGDSS--GDEKYAFMSDTTDITSSESEY 2551
Lambda K H
0.317 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4665550176
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40