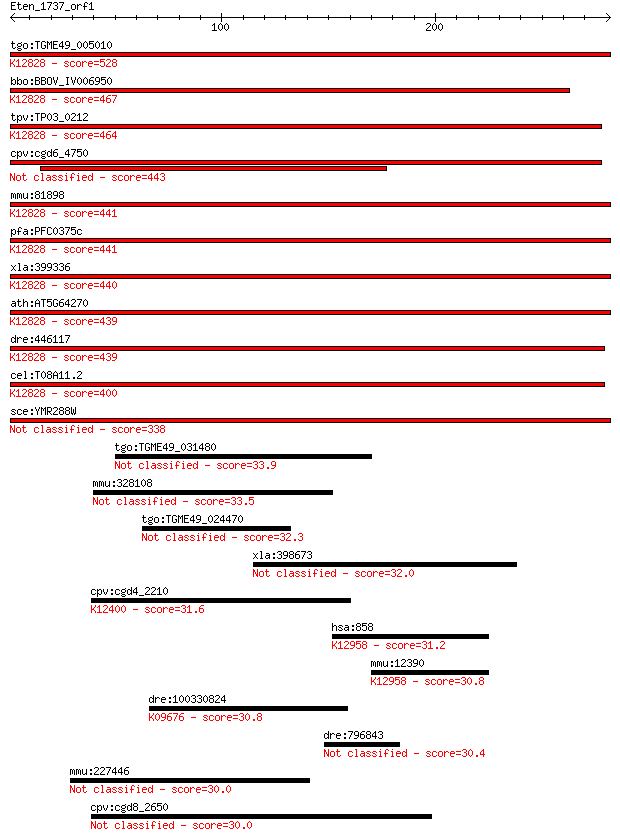
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1737_orf1
Length=281
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_005010 splicing factor 3B subunit 1, putative (EC:5... 528 8e-150
bbo:BBOV_IV006950 23.m06336; splicing factor; K12828 splicing ... 467 3e-131
tpv:TP03_0212 splicing factor 3B subunit 1; K12828 splicing fa... 464 1e-130
cpv:cgd6_4750 splicing factor 3B subunit1-like HEAT repeat con... 443 3e-124
mmu:81898 Sf3b1, 155kDa, 2810001M05Rik, AA409119, Prp10, SAP15... 441 2e-123
pfa:PFC0375c U2 snRNP spliceosome subunit, putative; K12828 sp... 441 2e-123
xla:399336 sf3b1, MGC115390, hsh155, prp10, prpf10, sap155; sp... 440 3e-123
ath:AT5G64270 splicing factor, putative; K12828 splicing facto... 439 4e-123
dre:446117 sf3b1, wu:fb99f09; splicing factor 3b, subunit 1; K... 439 5e-123
cel:T08A11.2 hypothetical protein; K12828 splicing factor 3B s... 400 3e-111
sce:YMR288W HSH155; Hsh155p 338 1e-92
tgo:TGME49_031480 translational activator, putative 33.9 0.78
mmu:328108 Fam179b, A430041B07Rik, BC062107, C130034K06, KIAA0... 33.5 0.89
tgo:TGME49_024470 hypothetical protein 32.3 2.0
xla:398673 ermp1, MGC86451; endoplasmic reticulum metallopepti... 32.0 2.6
cpv:cgd4_2210 hypothetical protein ; K12400 AP-4 complex subun... 31.6 3.1
hsa:858 CAV2, CAV, MGC12294; caveolin 2; K12958 caveolin 2 31.2
mmu:12390 Cav2, AI447843; caveolin 2; K12958 caveolin 2 30.8
dre:100330824 galactose-3-O-sulfotransferase 3-like; K09676 ga... 30.8 5.6
dre:796843 deformed epidermal autoregulatory factor 1-like 30.4 8.4
mmu:227446 2310035C23Rik, 6430401N10, Kiaa1468, mKIAA1468; RIK... 30.0 8.8
cpv:cgd8_2650 hypothetical protein 30.0 9.5
> tgo:TGME49_005010 splicing factor 3B subunit 1, putative (EC:5.5.1.4);
K12828 splicing factor 3B subunit 1
Length=1386
Score = 528 bits (1360), Expect = 8e-150, Method: Compositional matrix adjust.
Identities = 245/281 (87%), Positives = 265/281 (94%), Gaps = 0/281 (0%)
Query 1 LKNRHEKVQENLIDLIGRIADRGGDLVSPKEWDRICFDLLDLLRAQKKSIRRATVNTFGY 60
LKNRHEKVQEN+IDL+GRIADRGGDLVSPKEWDRICFDLLD+L+A KK+IRRATVNTFGY
Sbjct 1106 LKNRHEKVQENVIDLVGRIADRGGDLVSPKEWDRICFDLLDMLKASKKAIRRATVNTFGY 1165
Query 61 IARTIGPQDVLATLLNNLKMQERQLRLCTTIAIAIVAETCLPYSVLPALINEYRVPELNI 120
IARTIGPQDVLATLLNNLK+QERQLRLCTTIAIAIVAETCLPYSVLPAL+NEYRV ELN+
Sbjct 1166 IARTIGPQDVLATLLNNLKVQERQLRLCTTIAIAIVAETCLPYSVLPALMNEYRVQELNV 1225
Query 121 QNGVLKTLSFMFEYIGEMAKDYIYTVTPLLEDALMDRDLVHRQTAAWACKHLALGVHGLS 180
QNGVLKTLSFMFEYIGEMAKDYIYTV PLLEDALMDRDLVHRQTAAWA KHLALGVHGLS
Sbjct 1226 QNGVLKTLSFMFEYIGEMAKDYIYTVVPLLEDALMDRDLVHRQTAAWATKHLALGVHGLS 1285
Query 181 CEDALTHLFNYVWPNIFEVSAHMVQAFFDAVDGMRVAIGPGVVFRYVILGLFHPARKVRE 240
CEDAL HL N+VWPNIFE S H+VQAFFDAVDGMRV++G G+VFRYV+LGLFHPA+KVRE
Sbjct 1286 CEDALLHLMNFVWPNIFEKSPHLVQAFFDAVDGMRVSLGAGIVFRYVLLGLFHPAKKVRE 1345
Query 241 VYWRVYNNVYIGHQDAMVAFFPRLPDDGKHSFARHELELVI 281
VYWRVYNN+YIGHQD+MVAF+P LPDD K ++R EL VI
Sbjct 1346 VYWRVYNNLYIGHQDSMVAFYPPLPDDEKGCYSRDELLYVI 1386
> bbo:BBOV_IV006950 23.m06336; splicing factor; K12828 splicing
factor 3B subunit 1
Length=1147
Score = 467 bits (1201), Expect = 3e-131, Method: Compositional matrix adjust.
Identities = 211/262 (80%), Positives = 241/262 (91%), Gaps = 0/262 (0%)
Query 1 LKNRHEKVQENLIDLIGRIADRGGDLVSPKEWDRICFDLLDLLRAQKKSIRRATVNTFGY 60
LKNRHEKVQEN+I+L+GRIADRGGDLVSPKEWDRICFDLL+LL+A KK+IRRATVNTFGY
Sbjct 867 LKNRHEKVQENVIELVGRIADRGGDLVSPKEWDRICFDLLELLKANKKAIRRATVNTFGY 926
Query 61 IARTIGPQDVLATLLNNLKMQERQLRLCTTIAIAIVAETCLPYSVLPALINEYRVPELNI 120
IARTIGP DV+ATLLN+L++QERQLRLCTTIAIAIVAETCLPYSVLPAL+ EYRVPE+N+
Sbjct 927 IARTIGPNDVVATLLNHLRVQERQLRLCTTIAIAIVAETCLPYSVLPALMTEYRVPEINV 986
Query 121 QNGVLKTLSFMFEYIGEMAKDYIYTVTPLLEDALMDRDLVHRQTAAWACKHLALGVHGLS 180
Q GVLK L F+FEYIGEMAKDYIY +TPLLE+ALMDR+LVHRQTAAW CKHLALGV GL+
Sbjct 987 QTGVLKALCFLFEYIGEMAKDYIYAITPLLENALMDRNLVHRQTAAWTCKHLALGVAGLN 1046
Query 181 CEDALTHLFNYVWPNIFEVSAHMVQAFFDAVDGMRVAIGPGVVFRYVILGLFHPARKVRE 240
CEDAL HL NYVWPNIFE S H+ Q+ FDA+DG RVA+GPGV+F Y++ GLFHPA KVRE
Sbjct 1047 CEDALLHLLNYVWPNIFETSPHLTQSCFDAIDGFRVALGPGVIFNYILQGLFHPATKVRE 1106
Query 241 VYWRVYNNVYIGHQDAMVAFFP 262
VYWR+YNN+Y+G+QDA+V FP
Sbjct 1107 VYWRLYNNLYVGNQDALVPLFP 1128
> tpv:TP03_0212 splicing factor 3B subunit 1; K12828 splicing
factor 3B subunit 1
Length=1107
Score = 464 bits (1195), Expect = 1e-130, Method: Compositional matrix adjust.
Identities = 210/277 (75%), Positives = 245/277 (88%), Gaps = 0/277 (0%)
Query 1 LKNRHEKVQENLIDLIGRIADRGGDLVSPKEWDRICFDLLDLLRAQKKSIRRATVNTFGY 60
LKNRHEKVQEN+I+LIGRIADRGGDLVSPKEWDRICFDL+DLLRA KKSIRRATVNTFGY
Sbjct 827 LKNRHEKVQENVIELIGRIADRGGDLVSPKEWDRICFDLIDLLRANKKSIRRATVNTFGY 886
Query 61 IARTIGPQDVLATLLNNLKMQERQLRLCTTIAIAIVAETCLPYSVLPALINEYRVPELNI 120
IAR IGP DVL+TLLN+LK+QERQLR+CTTIAIAIVAETCLPYSVLPA++NEY++P+ NI
Sbjct 887 IARCIGPHDVLSTLLNHLKVQERQLRICTTIAIAIVAETCLPYSVLPAMMNEYKIPDQNI 946
Query 121 QNGVLKTLSFMFEYIGEMAKDYIYTVTPLLEDALMDRDLVHRQTAAWACKHLALGVHGLS 180
Q G+LK+L FMFEYIGEM+KDYIY++ PLLEDALM RDLVHRQTAAW CK+LALGV GL+
Sbjct 947 QTGILKSLCFMFEYIGEMSKDYIYSIVPLLEDALMCRDLVHRQTAAWTCKYLALGVFGLN 1006
Query 181 CEDALTHLFNYVWPNIFEVSAHMVQAFFDAVDGMRVAIGPGVVFRYVILGLFHPARKVRE 240
CEDAL HL NYVWPNIFE S H+ Q+ FDA+DG RV++GP V+F Y I GLFHPAR+VRE
Sbjct 1007 CEDALIHLLNYVWPNIFETSPHLTQSVFDALDGFRVSLGPSVIFYYTIQGLFHPARRVRE 1066
Query 241 VYWRVYNNVYIGHQDAMVAFFPRLPDDGKHSFARHEL 277
YWRVYNN+Y+GHQDA+V +P + + ++ +EL
Sbjct 1067 AYWRVYNNLYLGHQDALVPLYPLITEGVENKHQANEL 1103
> cpv:cgd6_4750 splicing factor 3B subunit1-like HEAT repeat containing
protein
Length=1031
Score = 443 bits (1140), Expect = 3e-124, Method: Compositional matrix adjust.
Identities = 197/277 (71%), Positives = 237/277 (85%), Gaps = 0/277 (0%)
Query 1 LKNRHEKVQENLIDLIGRIADRGGDLVSPKEWDRICFDLLDLLRAQKKSIRRATVNTFGY 60
LKNRHEKVQEN+I L+G A +GGD VSPKEWDRICFDLLD L+A KKSIRRA+V TFG+
Sbjct 751 LKNRHEKVQENIIQLLGCCAKKGGDFVSPKEWDRICFDLLDSLKANKKSIRRASVKTFGH 810
Query 61 IARTIGPQDVLATLLNNLKMQERQLRLCTTIAIAIVAETCLPYSVLPALINEYRVPELNI 120
IA+TIGPQDVL TLLNNL++QERQLR+CTTIAIAI++E C+PY+VLPA++NEYR+P+LN+
Sbjct 811 IAKTIGPQDVLVTLLNNLRVQERQLRVCTTIAIAIISEICMPYTVLPAIMNEYRIPDLNV 870
Query 121 QNGVLKTLSFMFEYIGEMAKDYIYTVTPLLEDALMDRDLVHRQTAAWACKHLALGVHGLS 180
QNGVLKTLSFMFEYIG M+KDYIY +TPLLE AL DRD VHRQTAAWACKHLALGV G
Sbjct 871 QNGVLKTLSFMFEYIGTMSKDYIYALTPLLEVALTDRDQVHRQTAAWACKHLALGVAGTG 930
Query 181 CEDALTHLFNYVWPNIFEVSAHMVQAFFDAVDGMRVAIGPGVVFRYVILGLFHPARKVRE 240
C DAL HL N++WPN+FE S H+VQA ++A+D RVA+GPGV+ Y++ GLFHPA+KVR
Sbjct 931 CNDALIHLLNFLWPNVFENSPHLVQAVYEALDAFRVALGPGVILNYLLQGLFHPAKKVRS 990
Query 241 VYWRVYNNVYIGHQDAMVAFFPRLPDDGKHSFARHEL 277
VYWR+YNN+YIG QD++V FFP +P G +F +E
Sbjct 991 VYWRIYNNLYIGSQDSLVPFFPPIPQIGNRNFDINEF 1027
Score = 30.8 bits (68), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 38/168 (22%), Positives = 64/168 (38%), Gaps = 47/168 (27%)
Query 15 LIGRIADRGGDLVSPKEW-DRICFDLLDLLRAQKKSIRRATVNTFGYIARTIGPQDVLAT 73
L I+D D +SP+E +R+ LL ++ +RR + AR GP +L
Sbjct 204 LFSNISD---DDLSPEEVNERLVLTLLLKIKNGAPILRRKAMKQIVETARDHGPGIILNH 260
Query 74 LL-----NNLKMQERQLRLCTTIAIAIVAETCLPYSVLPALINEYRVPELNIQNGVLKTL 128
LL + L+ QER + ++K L
Sbjct 261 LLPLLMQSTLEEQERHM--------------------------------------LVKAL 282
Query 129 SFMFEYIGEMAKDYIYTVTPLLEDALMDRDLVHRQTAAWACKHLALGV 176
+ + +GE K Y++ + ++E L+D+D RQ +LA V
Sbjct 283 DRILQRLGEKVKPYVHKILVVIEPMLIDQDYYARQEGREIISNLAKAV 330
> mmu:81898 Sf3b1, 155kDa, 2810001M05Rik, AA409119, Prp10, SAP155,
SF3b155, TA-8, Targ4; splicing factor 3b, subunit 1; K12828
splicing factor 3B subunit 1
Length=1304
Score = 441 bits (1133), Expect = 2e-123, Method: Compositional matrix adjust.
Identities = 197/281 (70%), Positives = 246/281 (87%), Gaps = 0/281 (0%)
Query 1 LKNRHEKVQENLIDLIGRIADRGGDLVSPKEWDRICFDLLDLLRAQKKSIRRATVNTFGY 60
LKNRHEKVQEN IDL+GRIADRG + VS +EW RICF+LL+LL+A KK+IRRATVNTFGY
Sbjct 1024 LKNRHEKVQENCIDLVGRIADRGAEYVSAREWMRICFELLELLKAHKKAIRRATVNTFGY 1083
Query 61 IARTIGPQDVLATLLNNLKMQERQLRLCTTIAIAIVAETCLPYSVLPALINEYRVPELNI 120
IA+ IGP DVLATLLNNLK+QERQ R+CTT+AIAIVAETC P++VLPAL+NEYRVPELN+
Sbjct 1084 IAKAIGPHDVLATLLNNLKVQERQNRVCTTVAIAIVAETCSPFTVLPALMNEYRVPELNV 1143
Query 121 QNGVLKTLSFMFEYIGEMAKDYIYTVTPLLEDALMDRDLVHRQTAAWACKHLALGVHGLS 180
QNGVLK+LSF+FEYIGEM KDYIY VTPLLEDALMDRDLVHRQTA+ +H++LGV+G
Sbjct 1144 QNGVLKSLSFLFEYIGEMGKDYIYAVTPLLEDALMDRDLVHRQTASAVVQHMSLGVYGFG 1203
Query 181 CEDALTHLFNYVWPNIFEVSAHMVQAFFDAVDGMRVAIGPGVVFRYVILGLFHPARKVRE 240
CED+L HL NYVWPN+FE S H++QA A++G+RVAIGP + +Y + GLFHPARKVR+
Sbjct 1204 CEDSLNHLLNYVWPNVFETSPHVIQAVMGALEGLRVAIGPCRMLQYCLQGLFHPARKVRD 1263
Query 241 VYWRVYNNVYIGHQDAMVAFFPRLPDDGKHSFARHELELVI 281
VYW++YN++YIG QDA++A +PR+ +D K+++ R+EL+ ++
Sbjct 1264 VYWKIYNSIYIGSQDALIAHYPRIYNDDKNTYIRYELDYIL 1304
> pfa:PFC0375c U2 snRNP spliceosome subunit, putative; K12828
splicing factor 3B subunit 1
Length=1386
Score = 441 bits (1133), Expect = 2e-123, Method: Compositional matrix adjust.
Identities = 197/281 (70%), Positives = 238/281 (84%), Gaps = 0/281 (0%)
Query 1 LKNRHEKVQENLIDLIGRIADRGGDLVSPKEWDRICFDLLDLLRAQKKSIRRATVNTFGY 60
LKNRHEKVQEN+IDLIG IAD+GGDLVSPKEWDRICFDL++LL++ KK IRRAT+ TFGY
Sbjct 1106 LKNRHEKVQENVIDLIGIIADKGGDLVSPKEWDRICFDLIELLKSNKKLIRRATIQTFGY 1165
Query 61 IARTIGPQDVLATLLNNLKMQERQLRLCTTIAIAIVAETCLPYSVLPALINEYRVPELNI 120
IARTIGP +VL LLNNLK+QERQLR+CTT+AIAIVA+TCLPYSVL AL+NEY+ ++N+
Sbjct 1166 IARTIGPFEVLTVLLNNLKVQERQLRVCTTVAIAIVADTCLPYSVLAALMNEYKTQDMNV 1225
Query 121 QNGVLKTLSFMFEYIGEMAKDYIYTVTPLLEDALMDRDLVHRQTAAWACKHLALGVHGLS 180
QNGVLK LSFMFEYIGE+AKDY+Y+V LLE ALMDRDLVHRQ A WACKHLALG GL+
Sbjct 1226 QNGVLKALSFMFEYIGEIAKDYVYSVVTLLEHALMDRDLVHRQIATWACKHLALGCFGLN 1285
Query 181 CEDALTHLFNYVWPNIFEVSAHMVQAFFDAVDGMRVAIGPGVVFRYVILGLFHPARKVRE 240
+DAL HL NYVWPNIFE S H++QA D++DG RVA+GP ++F+Y++ G+FHP+RKVRE
Sbjct 1286 RQDALIHLLNYVWPNIFETSPHLIQAVIDSIDGFRVALGPAIIFQYLVQGIFHPSRKVRE 1345
Query 241 VYWRVYNNVYIGHQDAMVAFFPRLPDDGKHSFARHELELVI 281
+YW++YNNVYIGHQD++V +P +F R EL I
Sbjct 1346 IYWKIYNNVYIGHQDSLVPIYPPFELLNDSTFVRDELRYTI 1386
> xla:399336 sf3b1, MGC115390, hsh155, prp10, prpf10, sap155;
splicing factor 3b, subunit 1, 155kDa; K12828 splicing factor
3B subunit 1
Length=1307
Score = 440 bits (1131), Expect = 3e-123, Method: Compositional matrix adjust.
Identities = 197/281 (70%), Positives = 245/281 (87%), Gaps = 0/281 (0%)
Query 1 LKNRHEKVQENLIDLIGRIADRGGDLVSPKEWDRICFDLLDLLRAQKKSIRRATVNTFGY 60
LKNRHEKVQEN IDL+GRIADRG + VS +EW RICF+LL+LL+A KK+IRRATVNTFGY
Sbjct 1027 LKNRHEKVQENCIDLVGRIADRGAEYVSAREWMRICFELLELLKAHKKAIRRATVNTFGY 1086
Query 61 IARTIGPQDVLATLLNNLKMQERQLRLCTTIAIAIVAETCLPYSVLPALINEYRVPELNI 120
IA+ IGP DVLATLLNNLK+QERQ R+CTT+AIAIVAETC P++VLPAL+NEYRVPELN+
Sbjct 1087 IAKAIGPHDVLATLLNNLKVQERQNRVCTTVAIAIVAETCSPFTVLPALMNEYRVPELNV 1146
Query 121 QNGVLKTLSFMFEYIGEMAKDYIYTVTPLLEDALMDRDLVHRQTAAWACKHLALGVHGLS 180
QNGVLK+LSF+FEYIGEM KDYIY VTPLLEDALMDRDLVHRQTA+ +H++LGV+G
Sbjct 1147 QNGVLKSLSFLFEYIGEMGKDYIYAVTPLLEDALMDRDLVHRQTASAVVQHMSLGVYGFG 1206
Query 181 CEDALTHLFNYVWPNIFEVSAHMVQAFFDAVDGMRVAIGPGVVFRYVILGLFHPARKVRE 240
CED+L HL NYVWPN+FE S H++QA A++G+RVAIGP + +Y + GLFHPARKVR+
Sbjct 1207 CEDSLNHLLNYVWPNVFETSPHVIQAVMGALEGLRVAIGPCRMVQYCLQGLFHPARKVRD 1266
Query 241 VYWRVYNNVYIGHQDAMVAFFPRLPDDGKHSFARHELELVI 281
VYW++YN++YIG QDA++A +PR+ +D K+++ R+EL+ +
Sbjct 1267 VYWKIYNSIYIGSQDALIAHYPRIYNDEKNTYIRYELDYTL 1307
> ath:AT5G64270 splicing factor, putative; K12828 splicing factor
3B subunit 1
Length=1269
Score = 439 bits (1130), Expect = 4e-123, Method: Compositional matrix adjust.
Identities = 200/281 (71%), Positives = 240/281 (85%), Gaps = 0/281 (0%)
Query 1 LKNRHEKVQENLIDLIGRIADRGGDLVSPKEWDRICFDLLDLLRAQKKSIRRATVNTFGY 60
LKNRHEKVQEN IDL+GRIADRG + V +EW RICF+LL++L+A KK IRRATVNTFGY
Sbjct 989 LKNRHEKVQENCIDLVGRIADRGAEFVPAREWMRICFELLEMLKAHKKGIRRATVNTFGY 1048
Query 61 IARTIGPQDVLATLLNNLKMQERQLRLCTTIAIAIVAETCLPYSVLPALINEYRVPELNI 120
IA+ IGPQDVLATLLNNLK+QERQ R+CTT+AIAIVAETC P++VLPAL+NEYRVPELN+
Sbjct 1049 IAKAIGPQDVLATLLNNLKVQERQNRVCTTVAIAIVAETCSPFTVLPALMNEYRVPELNV 1108
Query 121 QNGVLKTLSFMFEYIGEMAKDYIYTVTPLLEDALMDRDLVHRQTAAWACKHLALGVHGLS 180
QNGVLK+LSF+FEYIGEM KDYIY VTPLLEDALMDRDLVHRQTAA A KH+ALGV GL
Sbjct 1109 QNGVLKSLSFLFEYIGEMGKDYIYAVTPLLEDALMDRDLVHRQTAASAVKHMALGVAGLG 1168
Query 181 CEDALTHLFNYVWPNIFEVSAHMVQAFFDAVDGMRVAIGPGVVFRYVILGLFHPARKVRE 240
CEDAL HL N++WPNIFE S H++ A +A++GMRVA+G V+ Y + GLFHPARKVRE
Sbjct 1169 CEDALVHLLNFIWPNIFETSPHVINAVMEAIEGMRVALGAAVILNYCLQGLFHPARKVRE 1228
Query 241 VYWRVYNNVYIGHQDAMVAFFPRLPDDGKHSFARHELELVI 281
VYW++YN++YIG QD +VA +P L D+ + ++R EL + +
Sbjct 1229 VYWKIYNSLYIGAQDTLVAAYPVLEDEQNNVYSRPELTMFV 1269
> dre:446117 sf3b1, wu:fb99f09; splicing factor 3b, subunit 1;
K12828 splicing factor 3B subunit 1
Length=1315
Score = 439 bits (1130), Expect = 5e-123, Method: Compositional matrix adjust.
Identities = 198/278 (71%), Positives = 243/278 (87%), Gaps = 0/278 (0%)
Query 1 LKNRHEKVQENLIDLIGRIADRGGDLVSPKEWDRICFDLLDLLRAQKKSIRRATVNTFGY 60
LKNRHEKVQEN IDL+GRIADRG + VS +EW RICF+LL+LL+A KK+IRRATVNTFGY
Sbjct 1035 LKNRHEKVQENCIDLVGRIADRGAEYVSAREWMRICFELLELLKAHKKAIRRATVNTFGY 1094
Query 61 IARTIGPQDVLATLLNNLKMQERQLRLCTTIAIAIVAETCLPYSVLPALINEYRVPELNI 120
IA+ IGP DVLATLLNNLK+QERQ R+CTT+AIAIVAETC P++VLPAL+NEYRVPELN+
Sbjct 1095 IAKAIGPHDVLATLLNNLKVQERQNRVCTTVAIAIVAETCSPFTVLPALMNEYRVPELNV 1154
Query 121 QNGVLKTLSFMFEYIGEMAKDYIYTVTPLLEDALMDRDLVHRQTAAWACKHLALGVHGLS 180
QNGVLK+LSF+FEYIGEM KDYIY VTPLLEDALMDRDLVHRQTA+ +H++LGV+G
Sbjct 1155 QNGVLKSLSFLFEYIGEMGKDYIYAVTPLLEDALMDRDLVHRQTASAVVQHMSLGVYGFG 1214
Query 181 CEDALTHLFNYVWPNIFEVSAHMVQAFFDAVDGMRVAIGPGVVFRYVILGLFHPARKVRE 240
CED+L HL NYVWPN+FE S H++QA A++G+RVAIGP + +Y + GLFHPARKVR+
Sbjct 1215 CEDSLNHLLNYVWPNVFETSPHVIQAVMGALEGLRVAIGPCRMLQYCLQGLFHPARKVRD 1274
Query 241 VYWRVYNNVYIGHQDAMVAFFPRLPDDGKHSFARHELE 278
VYW++YN++YIG QDA++A +P + +D K+S+ R+ELE
Sbjct 1275 VYWKIYNSIYIGSQDALIAHYPLIFNDEKNSYVRYELE 1312
> cel:T08A11.2 hypothetical protein; K12828 splicing factor 3B
subunit 1
Length=1322
Score = 400 bits (1028), Expect = 3e-111, Method: Compositional matrix adjust.
Identities = 183/278 (65%), Positives = 228/278 (82%), Gaps = 0/278 (0%)
Query 1 LKNRHEKVQENLIDLIGRIADRGGDLVSPKEWDRICFDLLDLLRAQKKSIRRATVNTFGY 60
LKNRHEKVQEN IDL+G IADRG + VS +EW RICF+LL+LL+A KKSIRRA +NTFG+
Sbjct 1042 LKNRHEKVQENCIDLVGAIADRGSEFVSAREWMRICFELLELLKAHKKSIRRAAINTFGF 1101
Query 61 IARTIGPQDVLATLLNNLKMQERQLRLCTTIAIAIVAETCLPYSVLPALINEYRVPELNI 120
IA+ IGP DVLATLLNNLK+QERQLR+CTT+AIAIV+ETC P++VLPA++NEYRVPE+N+
Sbjct 1102 IAKAIGPHDVLATLLNNLKVQERQLRVCTTVAIAIVSETCAPFTVLPAIMNEYRVPEINV 1161
Query 121 QNGVLKTLSFMFEYIGEMAKDYIYTVTPLLEDALMDRDLVHRQTAAWACKHLALGVHGLS 180
QNGVLK LSFMFEYIGEMAKDYIY V PLL DALM+RD VHRQ A A HLA+GV+G
Sbjct 1162 QNGVLKALSFMFEYIGEMAKDYIYAVVPLLIDALMERDQVHRQIAVDAVAHLAIGVYGFG 1221
Query 181 CEDALTHLFNYVWPNIFEVSAHMVQAFFDAVDGMRVAIGPGVVFRYVILGLFHPARKVRE 240
CEDAL HL NYVWPN+ E S H++Q + A +GMRV++GP V +Y + L+HPARKVRE
Sbjct 1222 CEDALIHLLNYVWPNMLENSPHLIQRWVFACEGMRVSLGPIKVLQYCLQALWHPARKVRE 1281
Query 241 VYWRVYNNVYIGHQDAMVAFFPRLPDDGKHSFARHELE 278
W+V+NN+ +G DA++A +PR+ + + + R+EL+
Sbjct 1282 PVWKVFNNLILGSADALIAAYPRIENTPTNQYVRYELD 1319
> sce:YMR288W HSH155; Hsh155p
Length=971
Score = 338 bits (867), Expect = 1e-92, Method: Compositional matrix adjust.
Identities = 156/281 (55%), Positives = 198/281 (70%), Gaps = 2/281 (0%)
Query 1 LKNRHEKVQENLIDLIGRIADRGGDLVSPKEWDRICFDLLDLLRAQKKSIRRATVNTFGY 60
L+N+H KV+ N I +G I PKEW RICF+LL+LL++ K IRR+ TFG+
Sbjct 693 LRNKHRKVEVNTIKFVGLIGKLAPTYAPPKEWMRICFELLELLKSTNKEIRRSANATFGF 752
Query 61 IARTIGPQDVLATLLNNLKMQERQLRLCTTIAIAIVAETCLPYSVLPALINEYRVPELNI 120
IA IGP DVL LLNNLK+QERQLR+CT +AI IVA+ C PY+VLP ++NEY PE N+
Sbjct 753 IAEAIGPHDVLVALLNNLKVQERQLRVCTAVAIGIVAKVCGPYNVLPVIMNEYTTPETNV 812
Query 121 QNGVLKTLSFMFEYIGEMAKDYIYTVTPLLEDALMDRDLVHRQTAAWACKHLALGVHGLS 180
QNGVLK +SFMFEYIG M+KDYIY +TPLLEDAL DRDLVHRQTA+ HLAL G
Sbjct 813 QNGVLKAMSFMFEYIGNMSKDYIYFITPLLEDALTDRDLVHRQTASNVITHLALNCSGTG 872
Query 181 CEDALTHLFNYVWPNIFEVSAHMVQAFFDAVDGMRVAIGPGVVFRYVILGLFHPARKVRE 240
EDA HL N + PNIFE S H + + ++ + A+GPG+ Y+ GLFHPA+ VR+
Sbjct 873 HEDAFIHLMNLLIPNIFETSPHAIMRILEGLEALSQALGPGLFMNYIWAGLFHPAKNVRK 932
Query 241 VYWRVYNNVYIGHQDAMVAFFPRLPDDGKHSFARHELELVI 281
+WRVYNN+Y+ +QDAMV F+P PD+ + EL+LV+
Sbjct 933 AFWRVYNNMYVMYQDAMVPFYPVTPDNNEEYI--EELDLVL 971
> tgo:TGME49_031480 translational activator, putative
Length=3377
Score = 33.9 bits (76), Expect = 0.78, Method: Composition-based stats.
Identities = 33/130 (25%), Positives = 56/130 (43%), Gaps = 11/130 (8%)
Query 50 IRRATVNTFGYIARTIGPQ---DVLATLLNNLKMQERQL-RLCTTIAIAIVAETCLP--- 102
+R A FG IA+ +G + DVL+ L LK E + R ++ V P
Sbjct 2195 VRAAAARAFGTIAKGVGEEHLSDVLSWLFRTLKTSESSVERSGAAFGLSEVLVALGPDRL 2254
Query 103 YSVLPALI---NEYRVPELNIQNGVLKTLSFMFEYIGEMAKDYIYTVTPLLEDALMDRDL 159
+ LP ++ + + P +++ G L ++ E +DY+ V P+L L D
Sbjct 2255 KAFLPDILANATDQQAPP-DVREGYLGLFVYLPTAFRERFQDYVPEVLPVLLGGLADNAE 2313
Query 160 VHRQTAAWAC 169
R+ + AC
Sbjct 2314 PVREVSLRAC 2323
> mmu:328108 Fam179b, A430041B07Rik, BC062107, C130034K06, KIAA0423,
MGC189816, MGC189944, mKIAA0423; family with sequence
similarity 179, member B
Length=1776
Score = 33.5 bits (75), Expect = 0.89, Method: Composition-based stats.
Identities = 25/121 (20%), Positives = 54/121 (44%), Gaps = 13/121 (10%)
Query 40 LDLLRAQKKSIRRATVNTFGYIARTIGPQDVLATLLNNLKMQERQLR-----LCTTIAIA 94
+ +L K I++ + F + + +GPQ VL+ LL NLK + ++R +C +
Sbjct 440 VKVLADNKLVIKQEYMKIFLKLMKEVGPQRVLSLLLENLKHKHSRVREEVVNICICSLLT 499
Query 95 IVAETC----LPYSVLPALINEYRVPELNIQNGVLKTLSFMFEYIGEMAKDYIYTVTPLL 150
+E L + + PAL++ R ++ L+ + + +G + ++ +
Sbjct 500 YPSEDFDLPKLSFDLAPALVDSKR----RVRQAALEAFAVLASSMGSGKTNVLFKAVDTV 555
Query 151 E 151
E
Sbjct 556 E 556
> tgo:TGME49_024470 hypothetical protein
Length=743
Score = 32.3 bits (72), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 24/84 (28%), Positives = 39/84 (46%), Gaps = 15/84 (17%)
Query 63 RTIGPQD---VLATLLNNLKMQER------------QLRLCTTIAIAIVAETCLPYSVLP 107
+ +GP D +L L N ++ ER +L C IA + ETCL +SV
Sbjct 453 KKVGPTDDELLLNALFNPGRIFERLDDASESIFHISRLAACDFIADGVFEETCLQFSVSQ 512
Query 108 ALINEYRVPELNIQNGVLKTLSFM 131
++ ++ LN+Q VL +L +
Sbjct 513 GIVVQHYGEILNLQEPVLPSLGVL 536
> xla:398673 ermp1, MGC86451; endoplasmic reticulum metallopeptidase
1
Length=876
Score = 32.0 bits (71), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 34/129 (26%), Positives = 50/129 (38%), Gaps = 17/129 (13%)
Query 115 VPELN------IQNGVLKTLSFMFEYIGEMAKDYIYTVTPLLEDALMDRDLVHRQTAAWA 168
+PELN + L + F + K++ P + D LV R+ +W
Sbjct 699 IPELNETVRAPCDDAPLCGFPWYFPVHHLIRKNWYLPAPPAPINEHTDFKLVSREEMSWG 758
Query 169 CKHLALGVHGLSCEDALTHLFNYVWPNIFEVSAHMVQAFFDAVDGMRVAIGPGVVFRYVI 228
L V G S H+ YV PN + V + + DG+ VA G F +
Sbjct 759 ATRLFFEVKGPS------HMTVYVRPN-----TNTVLSSWSLGDGIPVASGETDYFIFYS 807
Query 229 LGLFHPARK 237
GL+ PA K
Sbjct 808 HGLYAPAWK 816
> cpv:cgd4_2210 hypothetical protein ; K12400 AP-4 complex subunit
epsilon-1
Length=910
Score = 31.6 bits (70), Expect = 3.1, Method: Composition-based stats.
Identities = 28/122 (22%), Positives = 54/122 (44%), Gaps = 17/122 (13%)
Query 39 LLDLLRAQKKSIRRATVNTFGYIARTIGPQDVLATLLNNLKMQERQLRLCTTIAIAIVAE 98
++D L + ++IRR T+ ++ Q V++ L+NNLK+ + C + I+
Sbjct 387 VVDCLEDKDETIRRCTLELLCNMSNPQNIQVVISKLINNLKIST-DIHFCKELVKNIL-- 443
Query 99 TCLPYSVLPALINEYRVPELNIQNGVLKTLSFMFEYIGE-MAKDYIYTVTPLLEDALMDR 157
L++E P N L T+ +FE GE + KD + + ++ +
Sbjct 444 ----------LLSEKFAPSY---NWYLNTMVSLFELSGEFVGKDKVNNIAQIIAEGPTGN 490
Query 158 DL 159
D+
Sbjct 491 DI 492
> hsa:858 CAV2, CAV, MGC12294; caveolin 2; K12958 caveolin 2
Length=162
Score = 31.2 bits (69), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 36/76 (47%), Gaps = 9/76 (11%)
Query 152 DALMDRDLVHRQTAAWACKHLALGVHGLSCEDALTHLFNYVW---PNIFEVSAHMVQAFF 208
D+ DRD HR + HL LG + E TH F+ VW +FE+S +++ F
Sbjct 35 DSDQDRD-PHRLNS-----HLKLGFEDVIAEPVTTHSFDKVWICSHALFEISKYVMYKFL 88
Query 209 DAVDGMRVAIGPGVVF 224
+ +A G++F
Sbjct 89 TVFLAIPLAFIAGILF 104
> mmu:12390 Cav2, AI447843; caveolin 2; K12958 caveolin 2
Length=162
Score = 30.8 bits (68), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 28/58 (48%), Gaps = 3/58 (5%)
Query 170 KHLALGVHGLSCEDALTHLFNYVW---PNIFEVSAHMVQAFFDAVDGMRVAIGPGVVF 224
HL LG L E TH F+ VW +FE+S +++ F + +A G++F
Sbjct 47 SHLKLGFEDLIAEPETTHSFDKVWICSHALFEISKYVMYKFLTVFLAIPLAFIAGILF 104
> dre:100330824 galactose-3-O-sulfotransferase 3-like; K09676
galactose-3-O-sulfotransferase 3 [EC:2.8.2.-]
Length=421
Score = 30.8 bits (68), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 26/94 (27%), Positives = 41/94 (43%), Gaps = 1/94 (1%)
Query 66 GPQDVLATLLNNLKMQERQLRLCTTIAIAIVAETCLPYSVLPALINEYRVPELNIQNGVL 125
P D++ + L + + R L TI I I+ E + L + N+Y + + NG L
Sbjct 105 SPPDIITSHLRFSRTELRHLMPNNTIYITILREPGAMFESLFSYYNQYCLSFKRVPNGSL 164
Query 126 KT-LSFMFEYIGEMAKDYIYTVTPLLEDALMDRD 158
+T L + Y KD +Y L D D+D
Sbjct 165 ETFLDEPWNYYRPDEKDSMYARNTLTFDLGGDKD 198
> dre:796843 deformed epidermal autoregulatory factor 1-like
Length=640
Score = 30.4 bits (67), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 19/36 (52%), Gaps = 1/36 (2%)
Query 148 PLLEDALMDRD-LVHRQTAAWACKHLALGVHGLSCE 182
P L D L RD L H +T + CK L +H L CE
Sbjct 417 PTLTDGLWKRDILCHGKTLNFLCKKKILQIHSLLCE 452
> mmu:227446 2310035C23Rik, 6430401N10, Kiaa1468, mKIAA1468; RIKEN
cDNA 2310035C23 gene
Length=1192
Score = 30.0 bits (66), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 34/123 (27%), Positives = 57/123 (46%), Gaps = 13/123 (10%)
Query 29 PKEWDRICFDLLDLLRAQKKSIRRATVNTFGYIARTIGPQDVLATLLN------NLKMQE 82
PKE D++ L +L++ R+ + AR +GP V A LL N K E
Sbjct 532 PKERDQLLHILFNLIKRPDDEQRQMILTGCVAFARHVGPTRVEAELLPQCWEQINHKYPE 591
Query 83 RQL---RLCTTIAIAIVAE--TCLPYSVLPALINEYRVPELNIQNGVLKTLSFMFEYIGE 137
R+L C +A + E + L S+L ++ E + +L ++ V+K+L + YI +
Sbjct 592 RRLLVAESCGALAPYLPKEIRSSLVLSMLQQMLMEDKA-DL-VREAVIKSLGIIMGYIDD 649
Query 138 MAK 140
K
Sbjct 650 PDK 652
> cpv:cgd8_2650 hypothetical protein
Length=3488
Score = 30.0 bits (66), Expect = 9.5, Method: Composition-based stats.
Identities = 34/159 (21%), Positives = 62/159 (38%), Gaps = 10/159 (6%)
Query 39 LLDLLRAQKKSIRRATVNTFGYIARTIGPQDVLATLLNNLKMQERQLRLCTTIAIAIVAE 98
LLDL+R+ + R + FG + + + +TL+ +L ++ + +
Sbjct 1113 LLDLIRSDAANKRSGAFSFFGKSLAVVQAEQLRSTLILSLGHSIGEVPILLFNSATETVR 1172
Query 99 TCLPYSVLPALINEYRVPELNIQNGVLKTLSFMFEYIGEMAKDYIYTVTPLLEDALMDRD 158
L VL + E + EL +Q LK + F+ + + T++ + L
Sbjct 1173 KIL--QVLETAVKEEK--ELQLQCSALKAIQVAFQALSSSESNQELTLSSRINSVL---- 1224
Query 159 LVHRQTAAWACKHLALGVHGLSCEDALTHLFNYVWPNIF 197
R + L G++GL C D LF+ IF
Sbjct 1225 --SRFHSGEGNTKLKKGIYGLDCGDISLELFSSFRDRIF 1261
Lambda K H
0.327 0.142 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10848096440
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40