bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1733_orf3
Length=134
Score E
Sequences producing significant alignments: (Bits) Value
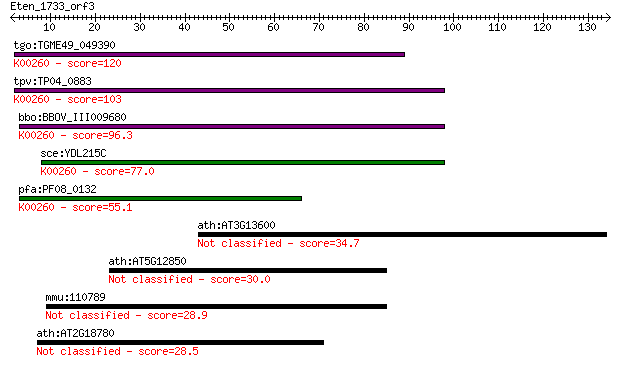
tgo:TGME49_049390 NAD-specific glutamate dehydrogenase, putati... 120 8e-28
tpv:TP04_0883 NAD-specific glutamate dehydrogenase (EC:1.4.1.2... 103 2e-22
bbo:BBOV_III009680 17.m07840; Glutamate/Leucine/Phenylalanine/... 96.3 3e-20
sce:YDL215C GDH2; Gdh2p (EC:1.4.1.2); K00260 glutamate dehydro... 77.0 1e-14
pfa:PF08_0132 glutamate dehydrogenase, putative (EC:1.4.1.2); ... 55.1 5e-08
ath:AT3G13600 calmodulin-binding family protein 34.7 0.081
ath:AT5G12850 zinc finger (CCCH-type) family protein 30.0 1.8
mmu:110789 Gpr98, Frings, Mass1, Mgr1, VLGR1; G protein-couple... 28.9 4.1
ath:AT2G18780 F-box family protein 28.5 6.6
> tgo:TGME49_049390 NAD-specific glutamate dehydrogenase, putative
(EC:1.4.1.2); K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1113
Score = 120 bits (302), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 55/87 (63%), Positives = 67/87 (77%), Gaps = 1/87 (1%)
Query 2 KTSSMLFDSSLLSPKGFKVPQDARDITLPDGTYVASGIEFRNNFHLSKFAVCDLFNPCGG 61
+TS+ML+D LLSP GFKV QD RD+ LPDGT VASG EFR FHL DLFNPCGG
Sbjct 818 QTSAMLYDEKLLSPMGFKVSQDDRDVVLPDGTAVASGFEFRGRFHLDPRGSADLFNPCGG 877
Query 62 RPASVNPRNVEQLFEQQTGLPKFKFII 88
RPASV P NV+++F+++ G P+FKFI+
Sbjct 878 RPASVTPFNVDKMFDEK-GKPRFKFIV 903
> tpv:TP04_0883 NAD-specific glutamate dehydrogenase (EC:1.4.1.2);
K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1178
Score = 103 bits (256), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 49/96 (51%), Positives = 68/96 (70%), Gaps = 1/96 (1%)
Query 2 KTSSMLFDSSLLSPKGFKVPQDARDITLPDGTYVASGIEFRNNFHLSKFAVCDLFNPCGG 61
KT SM +D LLS KGF VP++A ++ LPDG V +G +FR+ FHLS +A DLF PCGG
Sbjct 869 KTCSMGYDKRLLSSKGFMVPEEAMNVVLPDGFVVKNGYKFRDEFHLSSYAKADLFCPCGG 928
Query 62 RPASVNPRNVEQLFEQQTGLPKFKFIIILHCIYLLQ 97
RP+S+ P NV +LF+++ G +FKFI+ +Y+ Q
Sbjct 929 RPSSITPFNVNRLFDEK-GKCRFKFIVEGSNVYITQ 963
> bbo:BBOV_III009680 17.m07840; Glutamate/Leucine/Phenylalanine/Valine
dehydrogenase family protein (EC:1.4.1.2); K00260 glutamate
dehydrogenase [EC:1.4.1.2]
Length=1025
Score = 96.3 bits (238), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 48/95 (50%), Positives = 67/95 (70%), Gaps = 2/95 (2%)
Query 3 TSSMLFDSSLLSPKGFKVPQDARDITLPDGTYVASGIEFRNNFHLSKFAVCDLFNPCGGR 62
TS+M ++ +LLS KGFKVP+DA D+ LPDGT V G +FR+ FHL DLFNPCGGR
Sbjct 746 TSAMHYNEALLSDKGFKVPEDAVDMVLPDGTKVKRGHKFRDEFHLGA-CPSDLFNPCGGR 804
Query 63 PASVNPRNVEQLFEQQTGLPKFKFIIILHCIYLLQ 97
P+S+ P NV +LF+++ G +KFI+ +++ Q
Sbjct 805 PSSITPFNVNRLFDEK-GKCIYKFIVEGANVFITQ 838
> sce:YDL215C GDH2; Gdh2p (EC:1.4.1.2); K00260 glutamate dehydrogenase
[EC:1.4.1.2]
Length=1092
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 40/93 (43%), Positives = 54/93 (58%), Gaps = 3/93 (3%)
Query 8 FDSSLLSPKGFKVPQDARDITLPDGTYVASGIEFRNNFHLSKFAV---CDLFNPCGGRPA 64
FD+S LS GF V DA DI LP+GT VA+G FRN FH F D+F PCGGRP
Sbjct 806 FDTSKLSNNGFFVSVDAMDIMLPNGTIVANGTTFRNTFHTQIFKFVDHVDIFVPCGGRPN 865
Query 65 SVNPRNVEQLFEQQTGLPKFKFIIILHCIYLLQ 97
S+ N+ +++TG K +I+ +++ Q
Sbjct 866 SITLNNLHYFVDEKTGKCKIPYIVEGANLFITQ 898
> pfa:PF08_0132 glutamate dehydrogenase, putative (EC:1.4.1.2);
K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1397
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 25/63 (39%), Positives = 37/63 (58%), Gaps = 1/63 (1%)
Query 3 TSSMLFDSSLLSPKGFKVPQDARDITLPDGTYVASGIEFRNNFHLSKFAVCDLFNPCGGR 62
T L+D S GFK+ + ++ + G + +G++FRN F L+ C+LFNPCGGR
Sbjct 1039 TCCTLYDEKYFSKDGFKISIEDHNVDI-FGNKIRNGLDFRNTFFLNPLNKCELFNPCGGR 1097
Query 63 PAS 65
P S
Sbjct 1098 PHS 1100
> ath:AT3G13600 calmodulin-binding family protein
Length=605
Score = 34.7 bits (78), Expect = 0.081, Method: Compositional matrix adjust.
Identities = 28/92 (30%), Positives = 45/92 (48%), Gaps = 7/92 (7%)
Query 43 NNFHLSK-FAVCDLFNPCGGRPASVNPRNVEQLFEQQTGLPKFKFIIILHCIYLLQFYFM 101
N F ++K F+V D NP A++ + V + F + L ++ LL F +
Sbjct 88 NGFEIAKEFSVLDPRNP--KHEAAIKLQKVYKSFRTRRKLADCAVLVEQSWWKLLDFAEL 145
Query 102 EKTEETCSISIFIRESHQLALSRSSRDRRRGS 133
+++ SIS F E H+ A+SR SR R R +
Sbjct 146 KRS----SISFFDIEKHETAISRWSRARTRAA 173
> ath:AT5G12850 zinc finger (CCCH-type) family protein
Length=706
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 17/66 (25%), Positives = 31/66 (46%), Gaps = 11/66 (16%)
Query 23 DARDITLPDGTYVASGIEFRNNFHLSKFAVCDLFNP----CGGRPASVNPRNVEQLFEQQ 78
+ARDI + + + F + + D+ +P RP ++NP N+E+LF +
Sbjct 447 NARDIP-------SEQLSMLHEFEMQRQLAGDMHSPRFMNHSARPKTLNPSNLEELFSAE 499
Query 79 TGLPKF 84
P+F
Sbjct 500 VASPRF 505
> mmu:110789 Gpr98, Frings, Mass1, Mgr1, VLGR1; G protein-coupled
receptor 98
Length=6298
Score = 28.9 bits (63), Expect = 4.1, Method: Composition-based stats.
Identities = 25/78 (32%), Positives = 36/78 (46%), Gaps = 8/78 (10%)
Query 9 DSSLLSP-KGFKVPQDA-RDITLPDGTYVASGIEFRNNFHLSKFAVCDLFNPCGGRPASV 66
+S LSP KGF V ++ R TL + + E +F VC LFNP GG
Sbjct 3125 ESKDLSPSKGFIVLEEGVRSKTLRISAILDTEPEMDEHF------VCTLFNPTGGARLGA 3178
Query 67 NPRNVEQLFEQQTGLPKF 84
+ + + +F+ Q L F
Sbjct 3179 HVQTLITIFQNQAPLGLF 3196
> ath:AT2G18780 F-box family protein
Length=370
Score = 28.5 bits (62), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 20/72 (27%), Positives = 33/72 (45%), Gaps = 14/72 (19%)
Query 7 LFDSSLLSPKGFKVPQDARDITLPDGTYVASGIEFRNNFHLSKFAVCD--------LFNP 58
LF +LL+P + + + ++L G V + N FH F +C ++NP
Sbjct 78 LFSLNLLNPN--PIERLGQLVSLDGGAQV----DISNIFHCEGFLLCTTKDISRVVVWNP 131
Query 59 CGGRPASVNPRN 70
C G+ + PRN
Sbjct 132 CVGQTIWIKPRN 143
Lambda K H
0.324 0.138 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2231140792
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40