bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1724_orf2
Length=150
Score E
Sequences producing significant alignments: (Bits) Value
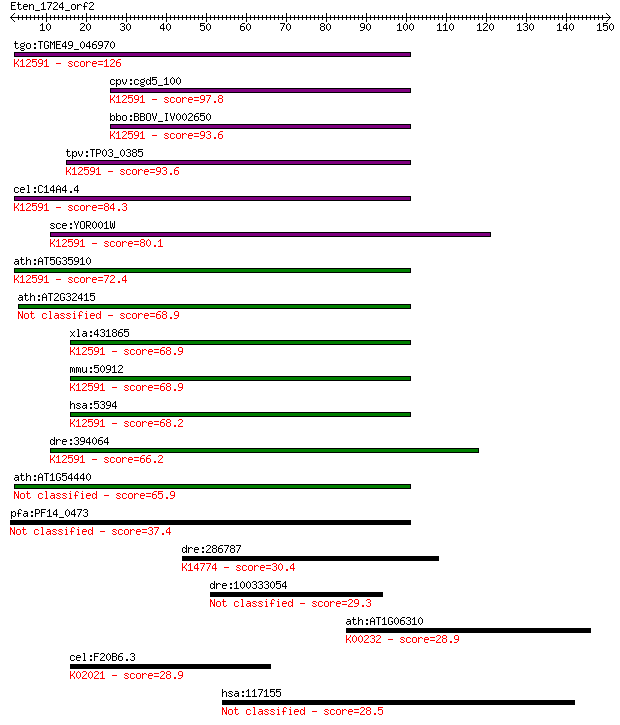
tgo:TGME49_046970 exosome component 10, putative ; K12591 exos... 126 3e-29
cpv:cgd5_100 RRPp/PMC2 like exosome 3'-5' exoribonuclease subu... 97.8 1e-20
bbo:BBOV_IV002650 21.m02990; exosome component 10; K12591 exos... 93.6 2e-19
tpv:TP03_0385 hypothetical protein; K12591 exosome complex exo... 93.6 2e-19
cel:C14A4.4 crn-3; Cell-death-Related Nuclease family member (... 84.3 1e-16
sce:YOR001W RRP6; Nuclear exosome exonuclease component; has 3... 80.1 2e-15
ath:AT5G35910 3'-5' exonuclease domain-containing protein / he... 72.4 5e-13
ath:AT2G32415 3'-5' exonuclease/ nucleic acid binding 68.9 6e-12
xla:431865 exosc10, MGC83774; exosome component 10; K12591 exo... 68.9 6e-12
mmu:50912 Exosc10, PM-Scl, PM/Scl-100, Pmscl2, RRP6, p2, p3, p... 68.9 6e-12
hsa:5394 EXOSC10, PM-Scl, PM/Scl-100, PMSCL, PMSCL2, RRP6, Rrp... 68.2 9e-12
dre:394064 MGC55695, Pmscl2, exosc10; zgc:55695; K12591 exosom... 66.2 3e-11
ath:AT1G54440 3'-5' exonuclease/ nucleic acid binding 65.9 5e-11
pfa:PF14_0473 Rrp6 homologue, putative 37.4 0.018
dre:286787 def, chunp6870, dj434o14.5, dj434o14.5l, fc51g08, f... 30.4 2.4
dre:100333054 cadherin 4-like 29.3 5.2
ath:AT1G06310 ACX6; ACX6 (ACYL-COA OXIDASE 6); FAD binding / a... 28.9 6.0
cel:F20B6.3 mrp-6; Multidrug Resistance Protein family member ... 28.9 6.6
hsa:117155 CATSPER2, MGC33346; cation channel, sperm associated 2 28.5
> tgo:TGME49_046970 exosome component 10, putative ; K12591 exosome
complex exonuclease RRP6 [EC:3.1.13.-]
Length=1353
Score = 126 bits (316), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 56/99 (56%), Positives = 75/99 (75%), Gaps = 5/99 (5%)
Query 2 PFMTVYRKQQLREMLDELCSGRHKVVAIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSL 61
P + + K++L+E++DEL SG H +VAID EHHS+ SY+GF CL+QLST KD +
Sbjct 460 PLVRIAEKEELQELVDELSSGAHSLVAIDLEHHSFHSYRGFTCLLQLST-----REKDYI 514
Query 62 VDPFSLFVHLTALNELTANPKILKILHGSASDVIWLRRE 100
+DPF+LF HL LN +TANPKILKI HG+ SD+IWL+R+
Sbjct 515 IDPFALFDHLHVLNTITANPKILKIFHGADSDIIWLQRD 553
> cpv:cgd5_100 RRPp/PMC2 like exosome 3'-5' exoribonuclease subunit
with an RNAseD domain and an HRDc domain ; K12591 exosome
complex exonuclease RRP6 [EC:3.1.13.-]
Length=957
Score = 97.8 bits (242), Expect = 1e-20, Method: Composition-based stats.
Identities = 45/75 (60%), Positives = 57/75 (76%), Gaps = 5/75 (6%)
Query 26 VVAIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSLVDPFSLFVHLTALNELTANPKILK 85
++AID EHHS +SYKGF+ LIQLST D ++DPF+LF + LNELTANPKILK
Sbjct 303 LLAIDVEHHSNQSYKGFVSLIQLST-----RTHDYIIDPFNLFNEIQMLNELTANPKILK 357
Query 86 ILHGSASDVIWLRRE 100
+LHGS D+IWL+R+
Sbjct 358 VLHGSDYDIIWLQRD 372
> bbo:BBOV_IV002650 21.m02990; exosome component 10; K12591 exosome
complex exonuclease RRP6 [EC:3.1.13.-]
Length=879
Score = 93.6 bits (231), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 37/75 (49%), Positives = 59/75 (78%), Gaps = 5/75 (6%)
Query 26 VVAIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSLVDPFSLFVHLTALNELTANPKILK 85
+VAID EHHS +SY+GF+CL+Q++ GA D ++DPFS+F + LN++T +P+ILK
Sbjct 337 IVAIDVEHHSTQSYRGFVCLVQIT-----GADDDWVIDPFSIFDEMWRLNDVTTDPRILK 391
Query 86 ILHGSASDVIWLRRE 100
++HG+ SD++WL+R+
Sbjct 392 VMHGAESDILWLQRD 406
> tpv:TP03_0385 hypothetical protein; K12591 exosome complex exonuclease
RRP6 [EC:3.1.13.-]
Length=996
Score = 93.6 bits (231), Expect = 2e-19, Method: Composition-based stats.
Identities = 42/86 (48%), Positives = 64/86 (74%), Gaps = 7/86 (8%)
Query 15 MLDELCSGRHKVVAIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSLVDPFSLFVHLTAL 74
MLD+L + R V+++D EHH E+Y+GFICL+QLST ++ ++DPF +F + L
Sbjct 484 MLDKLKNIR--VLSLDVEHHDTETYRGFICLVQLST-----PEENYIIDPFKIFGKMNKL 536
Query 75 NELTANPKILKILHGSASDVIWLRRE 100
N LT +PKILKI+HG+++DV+WL+R+
Sbjct 537 NRLTTDPKILKIMHGASNDVVWLQRD 562
> cel:C14A4.4 crn-3; Cell-death-Related Nuclease family member
(crn-3); K12591 exosome complex exonuclease RRP6 [EC:3.1.13.-]
Length=876
Score = 84.3 bits (207), Expect = 1e-16, Method: Composition-based stats.
Identities = 43/99 (43%), Positives = 60/99 (60%), Gaps = 7/99 (7%)
Query 2 PFMTVYRKQQLREMLDELCSGRHKVVAIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSL 61
P + K++L + L S K A+D EHH SY G CLIQ+ST +D +
Sbjct 277 PLTMIDTKEKLEALTKTLNS--VKEFAVDLEHHQMRSYLGLTCLIQIST-----RDEDFI 329
Query 62 VDPFSLFVHLTALNELTANPKILKILHGSASDVIWLRRE 100
+DPF ++ H+ LNE ANP+ILK+ HGS SDV+WL+R+
Sbjct 330 IDPFPIWDHVGMLNEPFANPRILKVFHGSDSDVLWLQRD 368
> sce:YOR001W RRP6; Nuclear exosome exonuclease component; has
3'-5' exonuclease activity; involved in RNA processing, maturation,
surveillance, degradation, tethering, and export; has
similarity to E. coli RNase D and to human PM-Sc1 100 (EXOSC10)
(EC:3.1.13.-); K12591 exosome complex exonuclease RRP6
[EC:3.1.13.-]
Length=733
Score = 80.1 bits (196), Expect = 2e-15, Method: Composition-based stats.
Identities = 44/110 (40%), Positives = 63/110 (57%), Gaps = 8/110 (7%)
Query 11 QLREMLDELCSGRHKVVAIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSLVDPFSLFVH 70
+L ML++L K +A+D EHH Y SY G +CL+Q+ST +D LVD L +
Sbjct 221 ELESMLEDL--KNTKEIAVDLEHHDYRSYYGIVCLMQIST-----RERDYLVDTLKLREN 273
Query 71 LTALNELTANPKILKILHGSASDVIWLRRESLGVMTTPSEPTSHQGRPDG 120
L LNE+ NP I+K+ HG+ D+IWL+R+ LG+ T H + G
Sbjct 274 LHILNEVFTNPSIVKVFHGAFMDIIWLQRD-LGLYVVGLFDTYHASKAIG 322
> ath:AT5G35910 3'-5' exonuclease domain-containing protein /
helicase and RNase D C-terminal domain-containing protein /
HRDC domain-containing protein; K12591 exosome complex exonuclease
RRP6 [EC:3.1.13.-]
Length=838
Score = 72.4 bits (176), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 37/100 (37%), Positives = 60/100 (60%), Gaps = 8/100 (8%)
Query 2 PFMTVYRKQQLREMLDELCSGRHKVVAIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSL 61
PF V + L+E++ +L S A+D EH+ Y S++G CL+Q+ST +D +
Sbjct 227 PFKFVQEVKDLKELVAKLRSVEE--FAVDLEHNQYRSFQGLTCLMQIST-----RTEDYI 279
Query 62 VDPFSLFVHL-TALNELTANPKILKILHGSASDVIWLRRE 100
VD F L +H+ L E+ +PK K++HG+ D+IWL+R+
Sbjct 280 VDTFKLRIHIGPYLREIFKDPKKKKVMHGADRDIIWLQRD 319
> ath:AT2G32415 3'-5' exonuclease/ nucleic acid binding
Length=891
Score = 68.9 bits (167), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 37/98 (37%), Positives = 60/98 (61%), Gaps = 7/98 (7%)
Query 3 FMTVYRKQQLREMLDELCSGRHKVVAIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSLV 62
++ V + QL+E+ + L + +V A+DTE HS S+ GF LIQ+ST +D LV
Sbjct 119 YVWVETESQLKELAEIL--AKEQVFAVDTEQHSLRSFLGFTALIQIST-----HEEDFLV 171
Query 63 DPFSLFVHLTALNELTANPKILKILHGSASDVIWLRRE 100
D +L ++ L + ++P I K+ HG+ +DVIWL+R+
Sbjct 172 DTIALHDVMSILRPVFSDPNICKVFHGADNDVIWLQRD 209
> xla:431865 exosc10, MGC83774; exosome component 10; K12591 exosome
complex exonuclease RRP6 [EC:3.1.13.-]
Length=883
Score = 68.9 bits (167), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 34/90 (37%), Positives = 54/90 (60%), Gaps = 10/90 (11%)
Query 16 LDELCSGRHKVV-----AIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSLVDPFSLFVH 70
L++L + K++ A+D EHHSY S+ G CL+Q+ST +D ++D L +
Sbjct 290 LEDLVALNEKLLQCAEFAVDLEHHSYRSFLGLTCLMQIST-----RTEDYIIDVLELRSN 344
Query 71 LTALNELTANPKILKILHGSASDVIWLRRE 100
L LNE NP I+K+ HG+ SD+ WL+++
Sbjct 345 LYILNESFTNPSIIKVFHGADSDIEWLQKD 374
> mmu:50912 Exosc10, PM-Scl, PM/Scl-100, Pmscl2, RRP6, p2, p3,
p4; exosome component 10; K12591 exosome complex exonuclease
RRP6 [EC:3.1.13.-]
Length=887
Score = 68.9 bits (167), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 35/90 (38%), Positives = 52/90 (57%), Gaps = 10/90 (11%)
Query 16 LDELCSGRHKVV-----AIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSLVDPFSLFVH 70
LDEL K++ A+D EHHSY S+ G CL+Q+ST +D +VD L
Sbjct 294 LDELVELNEKLLGCQEFAVDLEHHSYRSFLGLTCLMQIST-----RTEDFIVDTLELRSD 348
Query 71 LTALNELTANPKILKILHGSASDVIWLRRE 100
+ LNE +P I+K+ HG+ SD+ WL+++
Sbjct 349 MYILNESLTDPAIVKVFHGADSDIEWLQKD 378
> hsa:5394 EXOSC10, PM-Scl, PM/Scl-100, PMSCL, PMSCL2, RRP6, Rrp6p,
p2, p3, p4; exosome component 10; K12591 exosome complex
exonuclease RRP6 [EC:3.1.13.-]
Length=885
Score = 68.2 bits (165), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 34/90 (37%), Positives = 52/90 (57%), Gaps = 10/90 (11%)
Query 16 LDELCSGRHKVV-----AIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSLVDPFSLFVH 70
LDEL K++ A+D EHHSY S+ G CL+Q+ST +D ++D L
Sbjct 294 LDELVELNEKLLNCQEFAVDLEHHSYRSFLGLTCLMQIST-----RTEDFIIDTLELRSD 348
Query 71 LTALNELTANPKILKILHGSASDVIWLRRE 100
+ LNE +P I+K+ HG+ SD+ WL+++
Sbjct 349 MYILNESLTDPAIVKVFHGADSDIEWLQKD 378
> dre:394064 MGC55695, Pmscl2, exosc10; zgc:55695; K12591 exosome
complex exonuclease RRP6 [EC:3.1.13.-]
Length=899
Score = 66.2 bits (160), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 37/112 (33%), Positives = 58/112 (51%), Gaps = 11/112 (9%)
Query 11 QLREMLDELCSGRHKVV-----AIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSLVDPF 65
Q L++L + K+ A+D EHHSY S+ G CL+Q+ST +D ++D
Sbjct 287 QFISTLEDLVALNEKLAKTTEFAVDLEHHSYRSFLGITCLMQIST-----REEDFIIDTL 341
Query 66 SLFVHLTALNELTANPKILKILHGSASDVIWLRRESLGVMTTPSEPTSHQGR 117
L + LNE +P I+K+ HG+ SD+ WL+++ G+ T H R
Sbjct 342 ELRSEMYILNETFTDPAIVKVFHGADSDIEWLQKD-FGLYVVNMFDTHHAAR 392
> ath:AT1G54440 3'-5' exonuclease/ nucleic acid binding
Length=637
Score = 65.9 bits (159), Expect = 5e-11, Method: Composition-based stats.
Identities = 36/100 (36%), Positives = 57/100 (57%), Gaps = 8/100 (8%)
Query 2 PFMTVYRKQQLREMLDELCSGRHKVVAIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSL 61
PF V + L ++ L S A+D EH+ Y +++G CL+Q+ST +D +
Sbjct 114 PFKLVEEVKDLEDLAAALQSVEE--FAVDLEHNQYRTFQGLTCLMQIST-----RTEDYI 166
Query 62 VDPFSLFVHLTA-LNELTANPKILKILHGSASDVIWLRRE 100
VD F L+ H+ L EL +PK K++HG+ D+IWL+R+
Sbjct 167 VDIFKLWDHIGPYLRELFKDPKKKKVIHGADRDIIWLQRD 206
> pfa:PF14_0473 Rrp6 homologue, putative
Length=1136
Score = 37.4 bits (85), Expect = 0.018, Method: Composition-based stats.
Identities = 23/101 (22%), Positives = 53/101 (52%), Gaps = 6/101 (5%)
Query 1 KPFMTVYRKQQLREMLDEL-CSGRHKVVAIDTEHHSYESYKGFICLIQLSTCGSAGAAKD 59
K + + K L M++ + + K ++I + + +Y+GF +I + T +
Sbjct 537 KAYKIIDNKNDLINMINNIKLNYEEKKISIMIKVNYKRTYRGFTSIIMIGT-----NNMN 591
Query 60 SLVDPFSLFVHLTALNELTANPKILKILHGSASDVIWLRRE 100
++D F++F L +N++T +P ILKI + + + + L+++
Sbjct 592 YIIDVFNMFEDLYIINDITTDPNILKITYNAPNIINQLQKD 632
> dre:286787 def, chunp6870, dj434o14.5, dj434o14.5l, fc51g08,
fi05f05, wu:fc51g08, wu:fi05f05; digestive-organ expansion
factor; K14774 U3 small nucleolar RNA-associated protein 25
Length=753
Score = 30.4 bits (67), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 33/64 (51%), Gaps = 8/64 (12%)
Query 44 CLIQLSTCGSAGAAKDSLVDPFSLFVHLTALNELTANPKILKILHGSASDVIWLRRESLG 103
C+ L + G A + + P +H T L AN +L + G+A+DV L++E LG
Sbjct 230 CMSPLERFPAIGQASSAPLPP----IHKT----LEANWHLLNLPFGAATDVTELQKEMLG 281
Query 104 VMTT 107
+M T
Sbjct 282 LMGT 285
> dre:100333054 cadherin 4-like
Length=645
Score = 29.3 bits (64), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 22/44 (50%), Gaps = 1/44 (2%)
Query 51 CGSAGAAKDSLVDPFSLFVHLTALN-ELTANPKILKILHGSASD 93
CG+AGAA D PF HL A N E K + +LH S +
Sbjct 512 CGAAGAAGDFKAIPFDAKEHLIAYNTEGQGEDKGMALLHMSKDE 555
> ath:AT1G06310 ACX6; ACX6 (ACYL-COA OXIDASE 6); FAD binding /
acyl-CoA dehydrogenase/ acyl-CoA oxidase/ electron carrier/
oxidoreductase/ oxidoreductase, acting on the CH-CH group of
donors (EC:1.3.3.6); K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=675
Score = 28.9 bits (63), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 19/66 (28%), Positives = 31/66 (46%), Gaps = 5/66 (7%)
Query 85 KILHGSASDVIWLRRESLGVMTTPSEPT----SHQGRPD-GNVQSELHNACGARHPHRIR 139
K+ GS DV+ L R +++ +P+ H R + G+V+ E+ CG PH +
Sbjct 588 KLPPGSVKDVLGLVRSMYALISLEEDPSLLRYGHLSRDNVGDVRKEVSKLCGELRPHALA 647
Query 140 QGCSAG 145
S G
Sbjct 648 LVASFG 653
> cel:F20B6.3 mrp-6; Multidrug Resistance Protein family member
(mrp-6); K02021 putative ABC transport system ATP-binding
protein
Length=1396
Score = 28.9 bits (63), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 26/50 (52%), Gaps = 1/50 (2%)
Query 16 LDELCSGRHKVVAIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSLVDPF 65
L E CSGR + +D +H S +G + +I +G +++L DPF
Sbjct 1184 LKEYCSGRVTIDGVDLDHISLNFRRGGMSIIPQEPVIFSGTVRENL-DPF 1232
> hsa:117155 CATSPER2, MGC33346; cation channel, sperm associated
2
Length=528
Score = 28.5 bits (62), Expect = 9.3, Method: Composition-based stats.
Identities = 25/92 (27%), Positives = 36/92 (39%), Gaps = 8/92 (8%)
Query 54 AGAAKDSLVDPFSLFVHLTALNELTANPKILKILHGSASDVIWLRRE----SLGVMTTPS 109
A A + L+D FSL HL L++ I ++L S + L + +
Sbjct 15 ADAIRSRLIDTFSLIEHLQGLSQAVPRHTIRELLDPSRQKKLVLGDQHQLVRFSIKPQRI 74
Query 110 EPTSHQGRPDGNVQSELHNACGARHPHRIRQG 141
E SH R + S LH C R P + G
Sbjct 75 EQISHAQR----LLSRLHVRCSQRPPLSLWAG 102
Lambda K H
0.319 0.134 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3134054208
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40