bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1707_orf3
Length=287
Score E
Sequences producing significant alignments: (Bits) Value
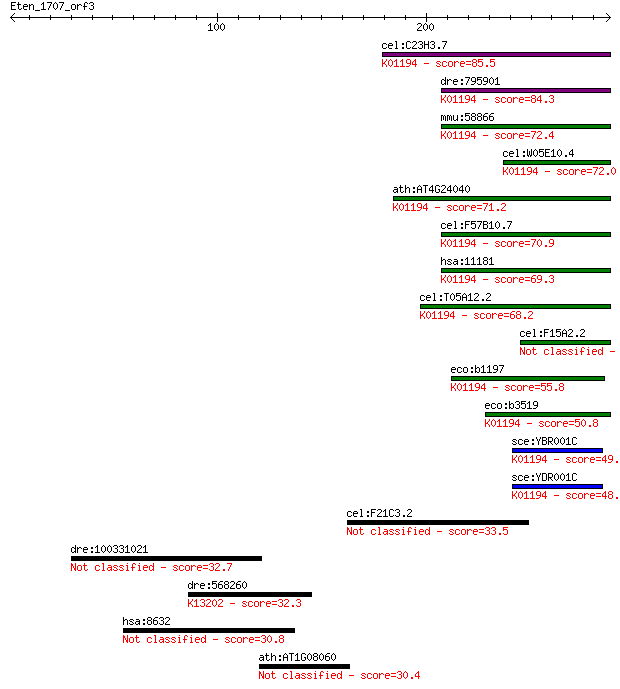
cel:C23H3.7 tre-5; TREhalase family member (tre-5); K01194 alp... 85.5 2e-16
dre:795901 treh, si:ch211-147p17.2; trehalase (brush-border me... 84.3 5e-16
mmu:58866 Treh, 2210412M19Rik; trehalase (brush-border membran... 72.4 2e-12
cel:W05E10.4 tre-3; TREhalase family member (tre-3); K01194 al... 72.0 2e-12
ath:AT4G24040 TRE1; TRE1 (TREHALASE 1); alpha,alpha-trehalase/... 71.2 4e-12
cel:F57B10.7 tre-1; TREhalase family member (tre-1); K01194 al... 70.9 5e-12
hsa:11181 TREH, MGC129621, TRE, TREA; trehalase (brush-border ... 69.3 2e-11
cel:T05A12.2 tre-2; TREhalase family member (tre-2); K01194 al... 68.2 3e-11
cel:F15A2.2 tre-4; TREhalase family member (tre-4) 63.2 1e-09
eco:b1197 treA, ECK1185, JW1186, osmA; periplasmic trehalase (... 55.8 2e-07
eco:b3519 treF, ECK3504, JW3487; cytoplasmic trehalase (EC:3.2... 50.8 5e-06
sce:YBR001C NTH2; Nth2p (EC:3.2.1.28); K01194 alpha,alpha-treh... 49.7 1e-05
sce:YDR001C NTH1; Neutral trehalase, degrades trehalose; requi... 48.1 4e-05
cel:F21C3.2 tyr-3; TYRosinase family member (tyr-3) 33.5 0.90
dre:100331021 hypothetical protein LOC100331021 32.7 1.6
dre:568260 si:dkey-40i22.3; K13202 target of EGR1 protein 1 32.3
hsa:8632 DNAH17, DNAHL1, DNEL2, FLJ40457, MGC132767, MGC138489... 30.8 5.3
ath:AT1G08060 MOM; MOM (MORPHEUS MOLECULE) 30.4 7.1
> cel:C23H3.7 tre-5; TREhalase family member (tre-5); K01194 alpha,alpha-trehalase
[EC:3.2.1.28]
Length=674
Score = 85.5 bits (210), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 43/109 (39%), Positives = 65/109 (59%), Gaps = 10/109 (9%)
Query 179 LPGEKGDTATVKNSEIMGENEFARRAVLLWAKLGRKTRRTVKTGSPIGSEASDNKIVSSS 238
LP + T + N + FA+R +W +L R+ + VK ++ S
Sbjct 158 LPDWRPITEQLANIKDASYQAFAQRLHFIWIQLCRQIKPEVK----------NDPSRFSL 207
Query 239 VHVPWGFIVPGGRFREMYYWDSYWIVRGLLHSGMISTARGIILNFADLV 287
++VP+ FI+PGGRFRE YYWD+YWI++GL+ S + STAR +ILNFA ++
Sbjct 208 IYVPYQFILPGGRFREFYYWDAYWILKGLIASELYSTARMMILNFAHII 256
> dre:795901 treh, si:ch211-147p17.2; trehalase (brush-border
membrane glycoprotein); K01194 alpha,alpha-trehalase [EC:3.2.1.28]
Length=577
Score = 84.3 bits (207), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 41/81 (50%), Positives = 51/81 (62%), Gaps = 10/81 (12%)
Query 207 LWAKLGRKTRRTVKTGSPIGSEASDNKIVSSSVHVPWGFIVPGGRFREMYYWDSYWIVRG 266
LW LGRK R V+ D+ + S ++ P +VPGGRFRE+YYWDSYW++ G
Sbjct 130 LWKSLGRKIRDDVR----------DHPELYSQIYTPHPVVVPGGRFRELYYWDSYWVING 179
Query 267 LLHSGMISTARGIILNFADLV 287
LL S M TARG+ILNF LV
Sbjct 180 LLLSEMTETARGMILNFVYLV 200
> mmu:58866 Treh, 2210412M19Rik; trehalase (brush-border membrane
glycoprotein) (EC:3.2.1.28); K01194 alpha,alpha-trehalase
[EC:3.2.1.28]
Length=576
Score = 72.4 bits (176), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 37/81 (45%), Positives = 48/81 (59%), Gaps = 10/81 (12%)
Query 207 LWAKLGRKTRRTVKTGSPIGSEASDNKIVSSSVHVPWGFIVPGGRFREMYYWDSYWIVRG 266
+W KLG+K + +E SS ++ FIVPGGRF E YYWDSYW++ G
Sbjct 131 IWKKLGKKMK----------AEVLSYPERSSLIYSKHPFIVPGGRFVEFYYWDSYWVMEG 180
Query 267 LLHSGMISTARGIILNFADLV 287
LL S M ST +G++ NF DLV
Sbjct 181 LLLSEMASTVKGMLQNFLDLV 201
> cel:W05E10.4 tre-3; TREhalase family member (tre-3); K01194
alpha,alpha-trehalase [EC:3.2.1.28]
Length=588
Score = 72.0 bits (175), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 30/51 (58%), Positives = 39/51 (76%), Gaps = 0/51 (0%)
Query 237 SSVHVPWGFIVPGGRFREMYYWDSYWIVRGLLHSGMISTARGIILNFADLV 287
S ++VP FIVPGGRFRE YYWD+YWI++GL+ S M +T R +I N A +V
Sbjct 142 SLLYVPNSFIVPGGRFREFYYWDAYWIIKGLIASDMYNTTRSMIRNLASMV 192
> ath:AT4G24040 TRE1; TRE1 (TREHALASE 1); alpha,alpha-trehalase/
trehalase (EC:3.2.1.28); K01194 alpha,alpha-trehalase [EC:3.2.1.28]
Length=626
Score = 71.2 bits (173), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 38/104 (36%), Positives = 60/104 (57%), Gaps = 13/104 (12%)
Query 184 GDTATVKNSEIMGENEFARRAVLLWAKLGRKTRRTVKTGSPIGSEASDNKIVSSSVHVPW 243
G + V+N E+ E+AR LW L + +V+ E++D + + +P
Sbjct 170 GFLSNVENEEV---REWAREVHGLWRNLSCRVSDSVR-------ESADRHTL---LPLPE 216
Query 244 GFIVPGGRFREMYYWDSYWIVRGLLHSGMISTARGIILNFADLV 287
I+PG RFRE+YYWDSYW+++GL+ S M +TA+G++ N LV
Sbjct 217 PVIIPGSRFREVYYWDSYWVIKGLMTSQMFTTAKGLVTNLMSLV 260
> cel:F57B10.7 tre-1; TREhalase family member (tre-1); K01194
alpha,alpha-trehalase [EC:3.2.1.28]
Length=567
Score = 70.9 bits (172), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 37/81 (45%), Positives = 48/81 (59%), Gaps = 10/81 (12%)
Query 207 LWAKLGRKTRRTVKTGSPIGSEASDNKIVSSSVHVPWGFIVPGGRFREMYYWDSYWIVRG 266
+W L RK R VK D+ S ++VP FI+PGGRF E YYWD++WI++G
Sbjct 111 IWKDLCRKVRDDVK-------HRQDH---YSLLYVPHPFIIPGGRFLEFYYWDTFWILKG 160
Query 267 LLHSGMISTARGIILNFADLV 287
LL S M TARG+I N +V
Sbjct 161 LLFSEMYETARGVIKNLGYMV 181
> hsa:11181 TREH, MGC129621, TRE, TREA; trehalase (brush-border
membrane glycoprotein) (EC:3.2.1.28); K01194 alpha,alpha-trehalase
[EC:3.2.1.28]
Length=583
Score = 69.3 bits (168), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 36/81 (44%), Positives = 46/81 (56%), Gaps = 10/81 (12%)
Query 207 LWAKLGRKTRRTVKTGSPIGSEASDNKIVSSSVHVPWGFIVPGGRFREMYYWDSYWIVRG 266
LW KLG+K + E + S ++ FIVPGGRF E YYWDSYW++ G
Sbjct 134 LWKKLGKKMK----------PEVLSHPERFSLIYSEHPFIVPGGRFVEFYYWDSYWVMEG 183
Query 267 LLHSGMISTARGIILNFADLV 287
LL S M T +G++ NF DLV
Sbjct 184 LLLSEMAETVKGMLQNFLDLV 204
> cel:T05A12.2 tre-2; TREhalase family member (tre-2); K01194
alpha,alpha-trehalase [EC:3.2.1.28]
Length=585
Score = 68.2 bits (165), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 38/94 (40%), Positives = 54/94 (57%), Gaps = 13/94 (13%)
Query 197 ENEFARRAVLL---WAKLGRKTRRTVKTGSPIGSEASDNKIVSSSVHVPWGFIVPGGRFR 253
+ ++ R A L W L RK + V+ N S + VP F+VPGGRFR
Sbjct 93 DEDYRRFAAALHAKWPTLYRKISKKVRV----------NPEKYSIIPVPNPFVVPGGRFR 142
Query 254 EMYYWDSYWIVRGLLHSGMISTARGIILNFADLV 287
EMYYWDS++ ++GL+ SGM++T +G+I N LV
Sbjct 143 EMYYWDSFFTIKGLIASGMLTTVKGMIENMIYLV 176
> cel:F15A2.2 tre-4; TREhalase family member (tre-4)
Length=635
Score = 63.2 bits (152), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 26/43 (60%), Positives = 34/43 (79%), Gaps = 0/43 (0%)
Query 245 FIVPGGRFREMYYWDSYWIVRGLLHSGMISTARGIILNFADLV 287
FIVPGGRF +YWD++WI++GLL S M T +GII NF++LV
Sbjct 178 FIVPGGRFDVYFYWDTFWIIKGLLVSRMFETTKGIINNFSNLV 220
> eco:b1197 treA, ECK1185, JW1186, osmA; periplasmic trehalase
(EC:3.2.1.28); K01194 alpha,alpha-trehalase [EC:3.2.1.28]
Length=565
Score = 55.8 bits (133), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 44/74 (59%), Gaps = 1/74 (1%)
Query 212 GRKTRRTVKTGSPIGSEASDN-KIVSSSVHVPWGFIVPGGRFREMYYWDSYWIVRGLLHS 270
G+ R + P+ + +++N + S + +P ++VPGGRFRE+YYWDSY+ + GL S
Sbjct 112 GQSLREHIDGLWPVLTRSTENTEKWDSLLPLPEPYVVPGGRFREVYYWDSYFTMLGLAES 171
Query 271 GMISTARGIILNFA 284
G ++ NFA
Sbjct 172 GHWDKVADMVANFA 185
> eco:b3519 treF, ECK3504, JW3487; cytoplasmic trehalase (EC:3.2.1.28);
K01194 alpha,alpha-trehalase [EC:3.2.1.28]
Length=549
Score = 50.8 bits (120), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 26/60 (43%), Positives = 36/60 (60%), Gaps = 0/60 (0%)
Query 228 EASDNKIVSSSVHVPWGFIVPGGRFREMYYWDSYWIVRGLLHSGMISTARGIILNFADLV 287
E D+ SS + +P +IVPGGRF E YYWDSY+ + GL SG + + NFA ++
Sbjct 145 EPQDHIPWSSLLALPQSYIVPGGRFSETYYWDSYFTMLGLAESGREDLLKCMADNFAWMI 204
> sce:YBR001C NTH2; Nth2p (EC:3.2.1.28); K01194 alpha,alpha-trehalase
[EC:3.2.1.28]
Length=780
Score = 49.7 bits (117), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 20/43 (46%), Positives = 29/43 (67%), Gaps = 0/43 (0%)
Query 241 VPWGFIVPGGRFREMYYWDSYWIVRGLLHSGMISTARGIILNF 283
V + + VPGGRF E+Y WDSY + GL+ S + ARG++ +F
Sbjct 321 VGYPYAVPGGRFNELYGWDSYLMALGLIESNKVDVARGMVEHF 363
> sce:YDR001C NTH1; Neutral trehalase, degrades trehalose; required
for thermotolerance and may mediate resistance to other
cellular stresses; may be phosphorylated by Cdc28p (EC:3.2.1.28);
K01194 alpha,alpha-trehalase [EC:3.2.1.28]
Length=751
Score = 48.1 bits (113), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 19/43 (44%), Positives = 28/43 (65%), Gaps = 0/43 (0%)
Query 241 VPWGFIVPGGRFREMYYWDSYWIVRGLLHSGMISTARGIILNF 283
+ + + VPGGRF E+Y WDSY + GLL + ARG++ +F
Sbjct 292 IGYPYAVPGGRFNELYGWDSYMMALGLLEANKTDVARGMVEHF 334
> cel:F21C3.2 tyr-3; TYRosinase family member (tyr-3)
Length=693
Score = 33.5 bits (75), Expect = 0.90, Method: Compositional matrix adjust.
Identities = 28/90 (31%), Positives = 43/90 (47%), Gaps = 12/90 (13%)
Query 162 PDISVCPTYTYGLPSPVLPGEKGDTATVKNSEIM-GENEFARRAVLLWA--KLGRKTRRT 218
P+ +V P YT+G P + G+ +T+T N I + F LLW + ++TR T
Sbjct 272 PNFNV-PEYTHGNPHIYVGGDMLETSTAANDPIFWMHHSFVD---LLWEMYRQSKQTRAT 327
Query 219 VKTGSPIGSEASDNKIVSSSVHVPWGFIVP 248
+T P +DN+ SS H F+ P
Sbjct 328 RETAYP-----ADNRQCSSEHHFRAAFMRP 352
> dre:100331021 hypothetical protein LOC100331021
Length=635
Score = 32.7 bits (73), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 26/99 (26%), Positives = 44/99 (44%), Gaps = 10/99 (10%)
Query 30 SLQQMRFGESITFV----DDIVCSDGPVFLAMRRLFNKVD----CKDLADLVMKLPVSAV 81
SL+++ F FV D++ DG VF +L D C+++ DLV P
Sbjct 162 SLEELGFSSMEAFVASLSKDLLVEDGVVFHKKTKLLTGEDFDKKCREIVDLVKSYPQGIP 221
Query 82 VARFANAPQEAEKRSLEILFQECFYSGFPDIIEWIPPDL 120
+A+ A+ A ++L +E +S D I + +L
Sbjct 222 LAQIASFYHTAYGKNLRK--KELGFSSLADFIHSLTQEL 258
> dre:568260 si:dkey-40i22.3; K13202 target of EGR1 protein 1
Length=483
Score = 32.3 bits (72), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 28/60 (46%), Gaps = 1/60 (1%)
Query 86 ANAPQEAEKRSLEILF-QECFYSGFPDIIEWIPPDLAESLPSWLTDTCPGVPEATRLTPS 144
AN P ++++F +CFY+ PD + DL++ PS + DT R T S
Sbjct 156 ANKPLVLHNGLIDLVFLYQCFYAHLPDRLGTFTADLSQMFPSGIYDTKYATEYELRFTAS 215
> hsa:8632 DNAH17, DNAHL1, DNEL2, FLJ40457, MGC132767, MGC138489;
dynein, axonemal, heavy chain 17
Length=4462
Score = 30.8 bits (68), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 45/90 (50%), Gaps = 13/90 (14%)
Query 55 LAMRRLFNKVDCKDLADLV-MKLPVSAVVARFANAPQEAEKRSLEILFQECFYSGFPDII 113
LA R+ K+D +D DL+ M+ VS++V NA +EAE+ + F+ Y ++
Sbjct 945 LAKDRMNYKMDLEDNTDLIEMREEVSSLVI---NAMKEAEE--YQDSFERYSYLWTDNLQ 999
Query 114 EWIPPDL-------AESLPSWLTDTCPGVP 136
E++ L AE L +W DT P P
Sbjct 1000 EFMKNFLIYGCAVTAEDLDTWTDDTIPKTP 1029
> ath:AT1G08060 MOM; MOM (MORPHEUS MOLECULE)
Length=2001
Score = 30.4 bits (67), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 120 LAESLPSWLTDTCPGVPEATRLTPSLRQKTPYNLTPSSPRVLP 162
LA S+P+ + PG+ ++R TPS + TP + T S R+ P
Sbjct 19 LAASIPASVEQETPGLRRSSRGTPSTKVITPASATRKSERLAP 61
Lambda K H
0.320 0.136 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 11230970432
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40