bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1687_orf2
Length=144
Score E
Sequences producing significant alignments: (Bits) Value
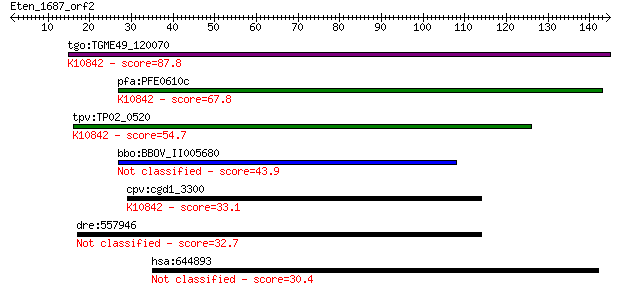
tgo:TGME49_120070 CDK-activating kinase assembly factor, putat... 87.8 1e-17
pfa:PFE0610c CDK-activating kinase assembly factor, putative; ... 67.8 1e-11
tpv:TP02_0520 hypothetical protein; K10842 CDK-activating kina... 54.7 1e-07
bbo:BBOV_II005680 18.m06472; hypothetical protein 43.9 2e-04
cpv:cgd1_3300 ring domain protein ; K10842 CDK-activating kina... 33.1 0.33
dre:557946 slain2, si:dkey-90m5.3; SLAIN motif family, member 2 32.7
hsa:644893 similar to hCG201263 30.4 2.0
> tgo:TGME49_120070 CDK-activating kinase assembly factor, putative
(EC:1.14.16.4); K10842 CDK-activating kinase assembly
factor MAT1
Length=279
Score = 87.8 bits (216), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 52/131 (39%), Positives = 74/131 (56%), Gaps = 3/131 (2%)
Query 15 QQNSEARAALRVRKIKEIVKKEGNFYEIVKLKPLGRNLKSSDESYVHALERQYSSLFQSA 74
Q+N E R KI +IV++EG FYE+VK +P E VH LERQY+ FQ
Sbjct 144 QENKEERKQREKEKIFQIVQREGIFYEVVKRRPALSRTTVDKEQLVHPLERQYAPYFQKE 203
Query 75 AAAAQVAAAAAAGSDGARPLDAAIREDQNIPRRKYLNINDFERAAAAAGYGPEVYRHRGR 134
VA A +G + ARPL+ +I++D ++PR +Y N FE+A A+GY P+ +G
Sbjct 204 ETTT-VAVRAESG-ETARPLNPSIKDDADVPRPRYKNREQFEKAELASGYAPQTVFAKGL 261
Query 135 QEL-SAILFAL 144
EL ++ F L
Sbjct 262 AELVGSVRFLL 272
> pfa:PFE0610c CDK-activating kinase assembly factor, putative;
K10842 CDK-activating kinase assembly factor MAT1
Length=260
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 39/116 (33%), Positives = 60/116 (51%), Gaps = 9/116 (7%)
Query 27 RKIKEIVKKEGNFYEIVKLKPLGRNLKSSDESYVHALERQYSSLFQSAAAAAQVAAAAAA 86
+KI EIVK+EGN YEI+K +P+ K +E+YVH+L ++ F A V
Sbjct 151 KKIHEIVKEEGNLYEIIKHRPIIN--KVHNETYVHSLIKENPKFFDEVKVANIVEVQ--- 205
Query 87 GSDGARPLDAAIREDQNIPRRKYLNINDFERAAAAAGYGPEVYRHRGRQELSAILF 142
+PL+ A + D +IP RKY + ++ +A A GY V R E + ++
Sbjct 206 ----PQPLNPAYKNDTDIPLRKYFSQDELYQADYAGGYDTNVVLKRCDIEFNKTIY 257
> tpv:TP02_0520 hypothetical protein; K10842 CDK-activating kinase
assembly factor MAT1
Length=245
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 37/111 (33%), Positives = 54/111 (48%), Gaps = 12/111 (10%)
Query 16 QNSEARAALRVRKIKEIVKKEGNFYEIV-KLKPLGRNLKSSDESYVHALERQYSSLFQSA 74
+N E + + IK IV+KEGNFYE+V K P NL DE +H L++ + F
Sbjct 140 KNFELHKSEEKKWIKSIVEKEGNFYELVDKGAPF--NLWKVDE-IIHPLKKSCPNYFTDE 196
Query 75 AAAAQVAAAAAAGSDGARPLDAAIREDQNIPRRKYLNINDFERAAAAAGYG 125
++PL+AAIR D +IPR+ + N + + A GY
Sbjct 197 TTVITTT--------DSKPLNAAIRNDTDIPRKVFTNRIELMESDVAGGYS 239
> bbo:BBOV_II005680 18.m06472; hypothetical protein
Length=175
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 30/82 (36%), Positives = 42/82 (51%), Gaps = 11/82 (13%)
Query 27 RKIKEIVKKEGNFYEIVKLKPLGRNLKSSDESYVHALERQYSSLFQSAAAAAQVAAAAAA 86
+++K IV E FYE+V K NL D+ VH L+R ++S F + A
Sbjct 99 KRVKHIVDTEKTFYEMVD-KSAPFNLWKVDD-LVHTLQRTHASYF--------IEEKALM 148
Query 87 GSDG-ARPLDAAIREDQNIPRR 107
S G PL+A+IR +IPRR
Sbjct 149 SSKGECVPLNASIRSTADIPRR 170
> cpv:cgd1_3300 ring domain protein ; K10842 CDK-activating kinase
assembly factor MAT1
Length=296
Score = 33.1 bits (74), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 28/90 (31%), Positives = 40/90 (44%), Gaps = 13/90 (14%)
Query 29 IKEIVKKEGNFYEIVKLKPLGRNLKSSDESYVHALERQY-----SSLFQSAAAAAQVAAA 83
I EIV+KEG FYE +L L +N VH L+ +Y + S
Sbjct 168 ILEIVQKEGTFYE--QLNQLSQNTTIDHLQIVHPLQNEYPEFFQNFNKNSNNQNNSNNQN 225
Query 84 AAAGSDGARPLDAAIREDQNIPRRKYLNIN 113
A GS+ P+ D+NI ++YL+ N
Sbjct 226 IALGSNIPNPI------DKNITCKEYLHQN 249
> dre:557946 slain2, si:dkey-90m5.3; SLAIN motif family, member
2
Length=567
Score = 32.7 bits (73), Expect = 0.38, Method: Composition-based stats.
Identities = 30/97 (30%), Positives = 45/97 (46%), Gaps = 5/97 (5%)
Query 17 NSEARAALRVRKIKEIVKKEGNFYEIVKLKPLGRNLKSSDESYVHALERQYSSLFQSAAA 76
NS A L VRK++++VKK E ++ + + S + R S+ F S A
Sbjct 5 NSNINADLEVRKLQDLVKKLEQQNEQLRSRSSLLSSSSPGGN-----TRPVSAGFDSPGA 59
Query 77 AAQVAAAAAAGSDGARPLDAAIREDQNIPRRKYLNIN 113
AA +A G G+R D A++ + PRR Y N
Sbjct 60 AAALAGFTGVGFGGSRGFDEALKNSRLSPRRSYHGSN 96
> hsa:644893 similar to hCG201263
Length=753
Score = 30.4 bits (67), Expect = 2.0, Method: Composition-based stats.
Identities = 30/111 (27%), Positives = 47/111 (42%), Gaps = 13/111 (11%)
Query 35 KEGNFYEIVKLKPLGRNLKSSDESYVHALERQYSSLFQSAAAAAQVAAAAAAGSDGARPL 94
++G+ YE V L+P+ R L+ S VH L+ + + +Q + + G+ P
Sbjct 555 EKGDSYEDV-LRPVHRFLQGS----VHELDSESEAEWQGLSVTLSCLKQTSPLKAGSAPS 609
Query 95 DAAIREDQNIPRRKYLNINDFERAAAAAGYGPEVYRHRGRQ----ELSAIL 141
E QN P +LN R A + P H+ + ELS IL
Sbjct 610 GTDTPELQNCPSTDFLNTLTTRRPA----FSPSQRTHKAARTQVPELSCIL 656
Lambda K H
0.315 0.127 0.340
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2814663556
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40