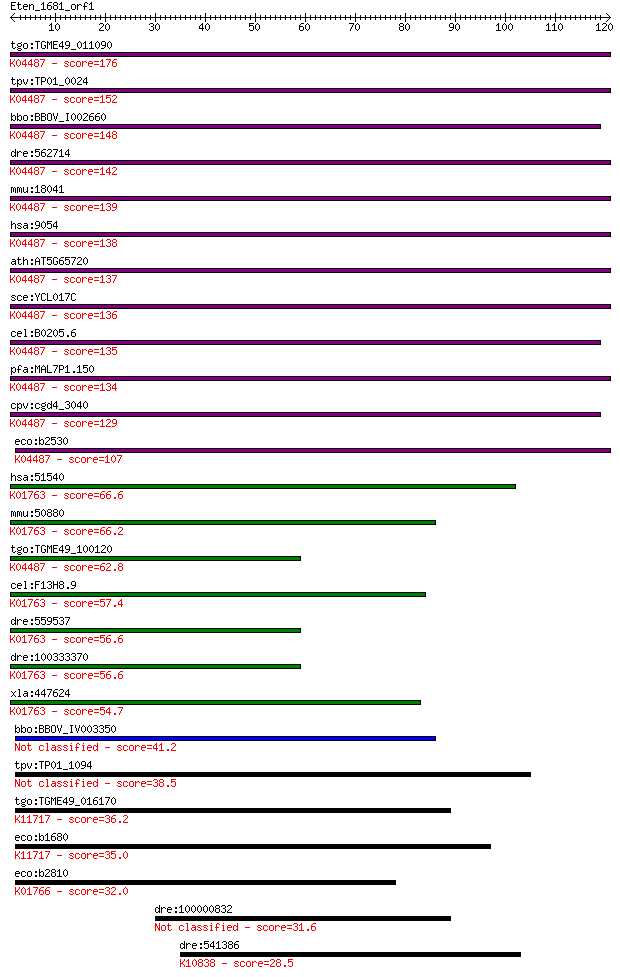
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1681_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_011090 cysteine desulfurase, putative (EC:2.8.1.7);... 176 1e-44
tpv:TP01_0024 cysteine desulfurase; K04487 cysteine desulfuras... 152 4e-37
bbo:BBOV_I002660 19.m02112; cysteine desulfurase (EC:2.8.1.7);... 148 4e-36
dre:562714 nfs1, fb50g03, wu:fb50g03; NFS1 nitrogen fixation 1... 142 2e-34
mmu:18041 Nfs1, AA987187, m-Nfs1, m-Nfsl; nitrogen fixation ge... 139 2e-33
hsa:9054 NFS1, IscS, NIFS; NFS1 nitrogen fixation 1 homolog (S... 138 5e-33
ath:AT5G65720 NFS1; NFS1; ATP binding / cysteine desulfurase/ ... 137 7e-33
sce:YCL017C NFS1, SPL1; Cysteine desulfurase involved in iron-... 136 1e-32
cel:B0205.6 hypothetical protein; K04487 cysteine desulfurase ... 135 3e-32
pfa:MAL7P1.150 cysteine desulfurase, putative (EC:4.4.1.-); K0... 134 5e-32
cpv:cgd4_3040 NifS-like protein; cysteine desulfurase ; K04487... 129 2e-30
eco:b2530 iscS, ECK2527, JW2514, nuvC, yfhO, yzzO; cysteine de... 107 1e-23
hsa:51540 SCLY, SCL, hSCL; selenocysteine lyase (EC:4.4.1.16);... 66.6 2e-11
mmu:50880 Scly, 9830169H08, A930015N15Rik, SCL, Scly1, Scly2; ... 66.2 3e-11
tgo:TGME49_100120 cysteine desulfurase, putative (EC:2.8.1.7);... 62.8 3e-10
cel:F13H8.9 hypothetical protein; K01763 selenocysteine lyase ... 57.4 1e-08
dre:559537 scly, zgc:171514; selenocysteine lyase (EC:4.4.1.16... 56.6 2e-08
dre:100333370 selenocysteine lyase-like; K01763 selenocysteine... 56.6 2e-08
xla:447624 scly, MGC85597; selenocysteine lyase (EC:4.4.1.16);... 54.7 7e-08
bbo:BBOV_IV003350 21.m02766; cysteine desulfurase (EC:2.8.1.7) 41.2 8e-04
tpv:TP01_1094 cysteine desulfurase 38.5 0.005
tgo:TGME49_016170 selenocysteine lyase, putative (EC:2.8.1.7);... 36.2 0.028
eco:b1680 sufS, csdB, ECK1676, JW1670, ynhB; cysteine desulfur... 35.0 0.054
eco:b2810 csdA, ECK2806, JW2781, ygdJ; cysteine sulfinate desu... 32.0 0.49
dre:100000832 probable RING-B-box-coiled coil protein-like 31.6 0.66
dre:541386 xpc, wu:fb12d05, wu:fb12d09, zgc:113032; xeroderma ... 28.5 5.5
> tgo:TGME49_011090 cysteine desulfurase, putative (EC:2.8.1.7);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=487
Score = 176 bits (446), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 84/120 (70%), Positives = 100/120 (83%), Gaps = 0/120 (0%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLAL 60
+S HKIYGPKG+GALFVRA+ P+VRL P+IDGGGQERG+RSGTL TPL VG G A LA
Sbjct 272 LSSHKIYGPKGIGALFVRAKNPRVRLQPLIDGGGQERGLRSGTLATPLCVGFGAACELAE 331
Query 61 ECMDSDRRHVERMARLLLHRLQQQLPHITVNGSLNRRYLGNLNISFSFVEGESLLMSIGD 120
+ M++DRRHV R+ARLLL +++Q+P I VNGSL RY GNLNISF+FVEGES+LMSI D
Sbjct 332 KEMENDRRHVSRLARLLLDSVREQIPDIEVNGSLTSRYPGNLNISFTFVEGESVLMSIRD 391
> tpv:TP01_0024 cysteine desulfurase; K04487 cysteine desulfurase
[EC:2.8.1.7]
Length=448
Score = 152 bits (383), Expect = 4e-37, Method: Composition-based stats.
Identities = 75/122 (61%), Positives = 95/122 (77%), Gaps = 4/122 (3%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLAL 60
+SGHKIYGPKGVGALFVR + P++RL PIIDGGGQERG+RSGTLPT L+VGLG AA +A
Sbjct 246 ISGHKIYGPKGVGALFVRTK-PRIRLQPIIDGGGQERGLRSGTLPTALVVGLGTAAKIAK 304
Query 61 ECMDSDRRHVERMARLLLHRLQQQLPHITVNGSLN--RRYLGNLNISFSFVEGESLLMSI 118
M+ D RH+E + L + L + H++VNGS+ +RY GNLN+SF F+EGESLLMS+
Sbjct 305 MEMERDHRHMENLFFKLYNGL-SSIDHVSVNGSIKPGQRYFGNLNMSFEFIEGESLLMSL 363
Query 119 GD 120
+
Sbjct 364 SN 365
> bbo:BBOV_I002660 19.m02112; cysteine desulfurase (EC:2.8.1.7);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=490
Score = 148 bits (373), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 74/120 (61%), Positives = 93/120 (77%), Gaps = 4/120 (3%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLAL 60
+SGHKIYGPKGVGAL+V R+P++RL PIIDGGGQERG+RSGTLP PL+VGLG+AA +A
Sbjct 288 ISGHKIYGPKGVGALYV-GRKPRIRLRPIIDGGGQERGLRSGTLPAPLVVGLGEAAKVAS 346
Query 61 ECMDSDRRHVERMARLLLHRLQQQLPHITVNGSLN--RRYLGNLNISFSFVEGESLLMSI 118
E M D H +R+ L+H + + H+ +NGS+ RY GNLN+SF FVEGESLLMS+
Sbjct 347 EEMHRDLEHAKRLWDKLVHGM-NDIGHVHINGSIQEGERYWGNLNVSFEFVEGESLLMSL 405
> dre:562714 nfs1, fb50g03, wu:fb50g03; NFS1 nitrogen fixation
1 (S. cerevisiae) (EC:2.8.1.7); K04487 cysteine desulfurase
[EC:2.8.1.7]
Length=451
Score = 142 bits (359), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 66/120 (55%), Positives = 92/120 (76%), Gaps = 1/120 (0%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLAL 60
+S HKIYGPKGVGALFVR RRP+VRL P+ GGGQERG+RSGT+PTPL VGLG A +A
Sbjct 248 ISAHKIYGPKGVGALFVR-RRPRVRLEPLQSGGGQERGLRSGTVPTPLAVGLGAACEIAQ 306
Query 61 ECMDSDRRHVERMARLLLHRLQQQLPHITVNGSLNRRYLGNLNISFSFVEGESLLMSIGD 120
+ ++ D + V +A L+ ++ ++P + +NG ++RY G +N+SF++VEGESLLM++ D
Sbjct 307 QELEYDHKRVSLLANRLVQKIMSEIPDVVMNGDPDQRYPGCINLSFAYVEGESLLMALKD 366
> mmu:18041 Nfs1, AA987187, m-Nfs1, m-Nfsl; nitrogen fixation
gene 1 (S. cerevisiae) (EC:2.8.1.7); K04487 cysteine desulfurase
[EC:2.8.1.7]
Length=459
Score = 139 bits (351), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 63/120 (52%), Positives = 91/120 (75%), Gaps = 1/120 (0%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLAL 60
+SGHK+YGPKGVGA+++R RRP+VR+ + GGGQERGMRSGT+PTPL+VGLG A LA
Sbjct 256 ISGHKLYGPKGVGAIYIR-RRPRVRVEALQSGGGQERGMRSGTVPTPLVVGLGAACELAQ 314
Query 61 ECMDSDRRHVERMARLLLHRLQQQLPHITVNGSLNRRYLGNLNISFSFVEGESLLMSIGD 120
+ M+ D + + ++A L+ + + LP + +NG + Y G +N+SF++VEGESLLM++ D
Sbjct 315 QEMEYDHKRISKLAERLVQNIMKNLPDVVMNGDPKQHYPGCINLSFAYVEGESLLMALKD 374
> hsa:9054 NFS1, IscS, NIFS; NFS1 nitrogen fixation 1 homolog
(S. cerevisiae) (EC:2.8.1.7); K04487 cysteine desulfurase [EC:2.8.1.7]
Length=406
Score = 138 bits (347), Expect = 5e-33, Method: Compositional matrix adjust.
Identities = 62/120 (51%), Positives = 90/120 (75%), Gaps = 1/120 (0%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLAL 60
+SGHKIYGPKGVGA+++R RRP+VR+ + GGGQERGMRSGT+PTPL+VGLG A +A
Sbjct 203 ISGHKIYGPKGVGAIYIR-RRPRVRVEALQSGGGQERGMRSGTVPTPLVVGLGAACEVAQ 261
Query 61 ECMDSDRRHVERMARLLLHRLQQQLPHITVNGSLNRRYLGNLNISFSFVEGESLLMSIGD 120
+ M+ D + + +++ L+ + + LP + +NG Y G +N+SF++VEGESLLM++ D
Sbjct 262 QEMEYDHKRISKLSERLIQNIMKSLPDVVMNGDPKHHYPGCINLSFAYVEGESLLMALKD 321
> ath:AT5G65720 NFS1; NFS1; ATP binding / cysteine desulfurase/
transaminase; K04487 cysteine desulfurase [EC:2.8.1.7]
Length=325
Score = 137 bits (346), Expect = 7e-33, Method: Compositional matrix adjust.
Identities = 65/120 (54%), Positives = 94/120 (78%), Gaps = 1/120 (0%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLAL 60
+S HKIYGPKGVGAL+VR RRP++RL P+++GGGQERG+RSGT T IVG G A LA+
Sbjct 122 MSAHKIYGPKGVGALYVR-RRPRIRLEPLMNGGGQERGLRSGTGATQQIVGFGAACELAM 180
Query 61 ECMDSDRRHVERMARLLLHRLQQQLPHITVNGSLNRRYLGNLNISFSFVEGESLLMSIGD 120
+ M+ D + ++ + LL+ ++++L + VNGS++ RY+GNLN+SF++VEGESLLM + +
Sbjct 181 KEMEYDEKWIKGLQERLLNGVREKLDGVVVNGSMDSRYVGNLNLSFAYVEGESLLMGLKE 240
> sce:YCL017C NFS1, SPL1; Cysteine desulfurase involved in iron-sulfur
cluster (Fe/S) biogenesis; required for the post-transcriptional
thio-modification of mitochondrial and cytoplasmic
tRNAs; essential protein located predominantly in mitochondria
(EC:2.8.1.7); K04487 cysteine desulfurase [EC:2.8.1.7]
Length=497
Score = 136 bits (343), Expect = 1e-32, Method: Composition-based stats.
Identities = 65/120 (54%), Positives = 91/120 (75%), Gaps = 2/120 (1%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLAL 60
+S HKIYGPKG+GA++VR RRP+VRL P++ GGGQERG+RSGTL PL+ G G+AA L
Sbjct 295 ISSHKIYGPKGIGAIYVR-RRPRVRLEPLLSGGGQERGLRSGTLAPPLVAGFGEAARLMK 353
Query 61 ECMDSDRRHVERMARLLLHRLQQQLPHITVNGSLNRRYLGNLNISFSFVEGESLLMSIGD 120
+ D+D+ H++R++ L+ L H T+NGS + RY G +N+SF++VEGESLLM++ D
Sbjct 354 KEFDNDQAHIKRLSDKLVKGLLSA-EHTTLNGSPDHRYPGCVNVSFAYVEGESLLMALRD 412
> cel:B0205.6 hypothetical protein; K04487 cysteine desulfurase
[EC:2.8.1.7]
Length=412
Score = 135 bits (341), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 64/118 (54%), Positives = 90/118 (76%), Gaps = 1/118 (0%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLAL 60
+S HKIYGPKG GAL+VR RRP+VR+ + GGGQERG+RSGT+ PL +GLG+AA +A
Sbjct 209 ISAHKIYGPKGAGALYVR-RRPRVRIEAQMSGGGQERGLRSGTVAAPLCIGLGEAAKIAD 267
Query 61 ECMDSDRRHVERMARLLLHRLQQQLPHITVNGSLNRRYLGNLNISFSFVEGESLLMSI 118
+ M D+ HVER++++L++ + +LPHI NG Y G +N+SF++VEGESLLM++
Sbjct 268 KEMAMDKAHVERLSQMLINGISDKLPHIIRNGDARHAYPGCVNLSFAYVEGESLLMAL 325
> pfa:MAL7P1.150 cysteine desulfurase, putative (EC:4.4.1.-);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=553
Score = 134 bits (338), Expect = 5e-32, Method: Composition-based stats.
Identities = 63/120 (52%), Positives = 84/120 (70%), Gaps = 0/120 (0%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLAL 60
+SGHK+YGPKG+GAL+++ ++P +RL +I GGGQERG+RSGTLPT LIVG G+AA +
Sbjct 346 MSGHKLYGPKGIGALYIKRKKPNIRLNALIHGGGQERGLRSGTLPTHLIVGFGEAAKVCS 405
Query 61 ECMDSDRRHVERMARLLLHRLQQQLPHITVNGSLNRRYLGNLNISFSFVEGESLLMSIGD 120
M+ D + V + L + L +I NG RY GN+NISF FVEGESLLMS+ +
Sbjct 406 LEMNRDEKKVRYFFNYVKDYLTKHLDYIVFNGCQINRYYGNMNISFLFVEGESLLMSLNE 465
> cpv:cgd4_3040 NifS-like protein; cysteine desulfurase ; K04487
cysteine desulfurase [EC:2.8.1.7]
Length=438
Score = 129 bits (324), Expect = 2e-30, Method: Composition-based stats.
Identities = 60/118 (50%), Positives = 83/118 (70%), Gaps = 1/118 (0%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLAL 60
+S HK+YGPKG+GA ++R++ P+ R+ P+I GGGQERGMRSGT+P PL VG G+A +A
Sbjct 235 LSAHKVYGPKGIGAFYIRSK-PRRRIKPLIFGGGQERGMRSGTMPVPLAVGFGEACKIAS 293
Query 61 ECMDSDRRHVERMARLLLHRLQQQLPHITVNGSLNRRYLGNLNISFSFVEGESLLMSI 118
M+SD HV+ + L + QLP + +NG R GNLN+SF+ VEGESL+M +
Sbjct 294 SEMNSDSIHVKSLYDKLYKGITTQLPDVELNGCGVNRMFGNLNLSFTGVEGESLMMKL 351
> eco:b2530 iscS, ECK2527, JW2514, nuvC, yfhO, yzzO; cysteine
desulfurase (tRNA sulfurtransferase), PLP-dependent (EC:2.8.1.7);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=404
Score = 107 bits (266), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 61/122 (50%), Positives = 85/122 (69%), Gaps = 8/122 (6%)
Query 2 SGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLALE 61
SGHKIYGPKG+GAL+VR R+P+VR+ + GGG ERGMRSGTLP IVG+G+A +A E
Sbjct 203 SGHKIYGPKGIGALYVR-RKPRVRIEAQMHGGGHERGMRSGTLPVHQIVGMGEAYRIAKE 261
Query 62 CMDSDRRHVERMARLLLHRLQ---QQLPHITVNGSLNRRYLGNLNISFSFVEGESLLMSI 118
M ++ +ER+ R L +RL + + + +NG L LN+SF++VEGESL+M++
Sbjct 262 EMATE---MERL-RGLRNRLWNGIKDIEEVYLNGDLEHGAPNILNVSFNYVEGESLIMAL 317
Query 119 GD 120
D
Sbjct 318 KD 319
> hsa:51540 SCLY, SCL, hSCL; selenocysteine lyase (EC:4.4.1.16);
K01763 selenocysteine lyase [EC:4.4.1.16]
Length=453
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 38/101 (37%), Positives = 57/101 (56%), Gaps = 3/101 (2%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLAL 60
+ GHK YGP+ +GAL++R L P++ GGGQER R GT TP+I GLGKAA L
Sbjct 263 IVGHKFYGPR-IGALYIRGLGEFTPLYPMLFGGGQERNFRPGTENTPMIAGLGKAAELVT 321
Query 61 ECMDSDRRHVERMARLLLHRLQQQLPHITVNGSLNRRYLGN 101
+ ++ H+ + L RL+ + ++ LN ++ G
Sbjct 322 QNCEAYEAHMRDVRDYLEERLEAEFGQKRIH--LNSQFPGT 360
> mmu:50880 Scly, 9830169H08, A930015N15Rik, SCL, Scly1, Scly2;
selenocysteine lyase (EC:4.4.1.16); K01763 selenocysteine
lyase [EC:4.4.1.16]
Length=432
Score = 66.2 bits (160), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 36/85 (42%), Positives = 49/85 (57%), Gaps = 1/85 (1%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLAL 60
+ GHK YGP+ +GAL+VR L P++ GGGQE R GT TP+I GLGKAA L
Sbjct 243 IVGHKFYGPR-IGALYVRGVGKLTPLYPMLFGGGQEWNFRPGTENTPMIAGLGKAADLVS 301
Query 61 ECMDSDRRHVERMARLLLHRLQQQL 85
E ++ H+ + L RL+ +
Sbjct 302 ENCETYEAHMRDIRDYLEERLEAEF 326
> tgo:TGME49_100120 cysteine desulfurase, putative (EC:2.8.1.7);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=851
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 32/58 (55%), Positives = 39/58 (67%), Gaps = 3/58 (5%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASL 58
++GHKIY PKGVGAL+V +R L P++ GGGQER +R GT P V LGKA L
Sbjct 404 IAGHKIYAPKGVGALYVGSR---ACLGPLLHGGGQERRLRGGTENVPYCVALGKACEL 458
> cel:F13H8.9 hypothetical protein; K01763 selenocysteine lyase
[EC:4.4.1.16]
Length=328
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 37/87 (42%), Positives = 49/87 (56%), Gaps = 7/87 (8%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLA- 59
V GHK YGP+ GAL + R+ P++ GG QE G RSGT TP+IVGLG+AA +
Sbjct 150 VVGHKFYGPRS-GALIFNPKSK--RIPPMLLGGNQESGWRSGTENTPMIVGLGEAAKVYN 206
Query 60 ---LECMDSDRRHVERMARLLLHRLQQ 83
L R++ + LLL RL+
Sbjct 207 EGFLNIESILRQNRDYFEELLLKRLRN 233
> dre:559537 scly, zgc:171514; selenocysteine lyase (EC:4.4.1.16);
K01763 selenocysteine lyase [EC:4.4.1.16]
Length=450
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 30/58 (51%), Positives = 35/58 (60%), Gaps = 1/58 (1%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASL 58
+ GHK YGP+ GALFV + P+ GGGQER R GT T +I GLGKAA L
Sbjct 260 IVGHKFYGPR-TGALFVNDPGKSTPVYPMFFGGGQERNFRPGTENTAMIAGLGKAAEL 316
> dre:100333370 selenocysteine lyase-like; K01763 selenocysteine
lyase [EC:4.4.1.16]
Length=450
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 30/58 (51%), Positives = 35/58 (60%), Gaps = 1/58 (1%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASL 58
+ GHK YGP+ GALFV + P+ GGGQER R GT T +I GLGKAA L
Sbjct 260 IVGHKFYGPR-TGALFVNDPGKSTPVYPMFFGGGQERNFRPGTENTAMIAGLGKAAEL 316
> xla:447624 scly, MGC85597; selenocysteine lyase (EC:4.4.1.16);
K01763 selenocysteine lyase [EC:4.4.1.16]
Length=426
Score = 54.7 bits (130), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 34/83 (40%), Positives = 50/83 (60%), Gaps = 3/83 (3%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASL-A 59
+ GHK YGP+ +GAL+V + L P++ GGG+E R GT TP+I GLG+AA L +
Sbjct 235 IVGHKFYGPR-IGALYVGGLGHQSPLLPMLYGGGREGNFRPGTENTPMIAGLGQAAELVS 293
Query 60 LECMDSDRRHVERMARLLLHRLQ 82
L C + H+ R+ L +L+
Sbjct 294 LHCA-AYEVHMRRIRDYLEQKLE 315
> bbo:BBOV_IV003350 21.m02766; cysteine desulfurase (EC:2.8.1.7)
Length=423
Score = 41.2 bits (95), Expect = 8e-04, Method: Composition-based stats.
Identities = 29/107 (27%), Positives = 48/107 (44%), Gaps = 24/107 (22%)
Query 2 SGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGM--------------RSGTLPTP 47
SGHK+YGP G+G L+ + + ++P GGG + + SGT P
Sbjct 272 SGHKVYGPTGIGFLYAKQEVIE-HMSPYKGGGGMVKNLDTSGYELEDIPHRFESGTPPVA 330
Query 48 LIVGLGKA---------ASLALECMDSDRRHVERMARLLLHRLQQQL 85
+GL KA +++ ++ + S+ + R+LL L QL
Sbjct 331 QAIGLSKALEFISHIGLSNVRMKGVLSNPHQIASHERMLLSHLDTQL 377
> tpv:TP01_1094 cysteine desulfurase
Length=469
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 31/118 (26%), Positives = 52/118 (44%), Gaps = 16/118 (13%)
Query 2 SGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGM--------------RSGTLPTP 47
S HK+YGP GVG L+ + R + L P GGG + + +GT P
Sbjct 284 SSHKMYGPTGVGFLYYKKRLLE-DLEPQKCGGGTVKDVTFESCDYFEPPFRFEAGTPPVA 342
Query 48 LIVGLGKAASLALEC-MDSDRRHVERMARLLLHRLQQQLPHITVNGSLNRRYLGNLNI 104
+GLG A +D R+H + + L R++ + + +L+R + + N+
Sbjct 343 QAIGLGAAVDFINSIGIDRIRKHDNMLLQYLYERMKDLVNLYNFDPTLDRIPIISFNV 400
> tgo:TGME49_016170 selenocysteine lyase, putative (EC:2.8.1.7);
K11717 cysteine desulfurase / selenocysteine lyase [EC:2.8.1.7
4.4.1.16]
Length=596
Score = 36.2 bits (82), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 30/103 (29%), Positives = 42/103 (40%), Gaps = 18/103 (17%)
Query 2 SGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQ---------------ERGMRSGTLPT 46
SGHK+YGP GVG F+ + +R P GGG+ +GT P
Sbjct 327 SGHKMYGPTGVG--FLYGKYELLRNMPPWKGGGEMIEFVDLCESTYANPPARFEAGTPPF 384
Query 47 PLIVGLGKAASLALEC-MDSDRRHVERMARLLLHRLQQQLPHI 88
++GLG A E + H R+ R L L + P +
Sbjct 385 LQVIGLGAAVDFIEEVGWPAIYSHDARLQRALHEVLASRFPEL 427
> eco:b1680 sufS, csdB, ECK1676, JW1670, ynhB; cysteine desulfurase,
stimulated by SufE; selenocysteine lyase, PLP-dependent
(EC:4.4.1.16); K11717 cysteine desulfurase / selenocysteine
lyase [EC:2.8.1.7 4.4.1.16]
Length=406
Score = 35.0 bits (79), Expect = 0.054, Method: Compositional matrix adjust.
Identities = 34/115 (29%), Positives = 51/115 (44%), Gaps = 26/115 (22%)
Query 2 SGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERG----------------MRSGTLP 45
SGHK+YGP G+G L+V + ++ P +GGG +GT
Sbjct 223 SGHKLYGPTGIGILYV--KEALLQEMPPWEGGGSMIATVSLSEGTTWTKAPWRFEAGTPN 280
Query 46 TPLIVGLGKAASLALECMDS-DRRHVERMARLLLHRLQQQL---PHITVNGSLNR 96
T I+GLG ALE + + ++ + L+H QL P +T+ G NR
Sbjct 281 TGGIIGLGA----ALEYVSALGLNNIAEYEQNLMHYALSQLESVPDLTLYGPQNR 331
> eco:b2810 csdA, ECK2806, JW2781, ygdJ; cysteine sulfinate desulfinase
(EC:4.4.1.-); K01766 cysteine sulfinate desulfinase
[EC:4.4.1.-]
Length=401
Score = 32.0 bits (71), Expect = 0.49, Method: Composition-based stats.
Identities = 27/87 (31%), Positives = 40/87 (45%), Gaps = 12/87 (13%)
Query 2 SGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQER-----GMRSGTLPTPLIVGLGKAA 56
SGHK+YGP G+G L+ ++ + ++P + GG G + + P L G A
Sbjct 219 SGHKLYGPTGIGVLYGKSELLEA-MSPWLGGGKMVHEVSFDGFTTQSAPWKLEAGTPNVA 277
Query 57 -----SLALECM-DSDRRHVERMARLL 77
S ALE + D D E +R L
Sbjct 278 GVIGLSAALEWLADYDINQAESWSRSL 304
> dre:100000832 probable RING-B-box-coiled coil protein-like
Length=604
Score = 31.6 bits (70), Expect = 0.66, Method: Composition-based stats.
Identities = 17/59 (28%), Positives = 29/59 (49%), Gaps = 0/59 (0%)
Query 30 IDGGGQERGMRSGTLPTPLIVGLGKAASLALECMDSDRRHVERMARLLLHRLQQQLPHI 88
I+ Q RS L LI L ++ + LE + +RR E A L++ L+Q++ +
Sbjct 274 INALAQHEAQRSMQLFGALISSLERSQAGLLEVTEINRRSAEHQAELMIRELEQEITEL 332
> dre:541386 xpc, wu:fb12d05, wu:fb12d09, zgc:113032; xeroderma
pigmentosum, complementation group C; K10838 xeroderma pigmentosum
group C-complementing protein
Length=879
Score = 28.5 bits (62), Expect = 5.5, Method: Composition-based stats.
Identities = 17/68 (25%), Positives = 35/68 (51%), Gaps = 2/68 (2%)
Query 35 QERGMRSGTLPTPLIVGLGKAASLALECMDSDRRHVERMARLLLHRLQQQLPHITVNGSL 94
+ R +R G P +++G + A M S++++V+ +A + ++ P I V+G +
Sbjct 629 EARTVRLGEEPYKMVLGFSNRSRKAR--MMSEQKNVKDLALFGTWQTEEYQPPIAVDGKV 686
Query 95 NRRYLGNL 102
R GN+
Sbjct 687 PRNEFGNV 694
Lambda K H
0.323 0.141 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2018002440
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40