bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1658_orf2
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
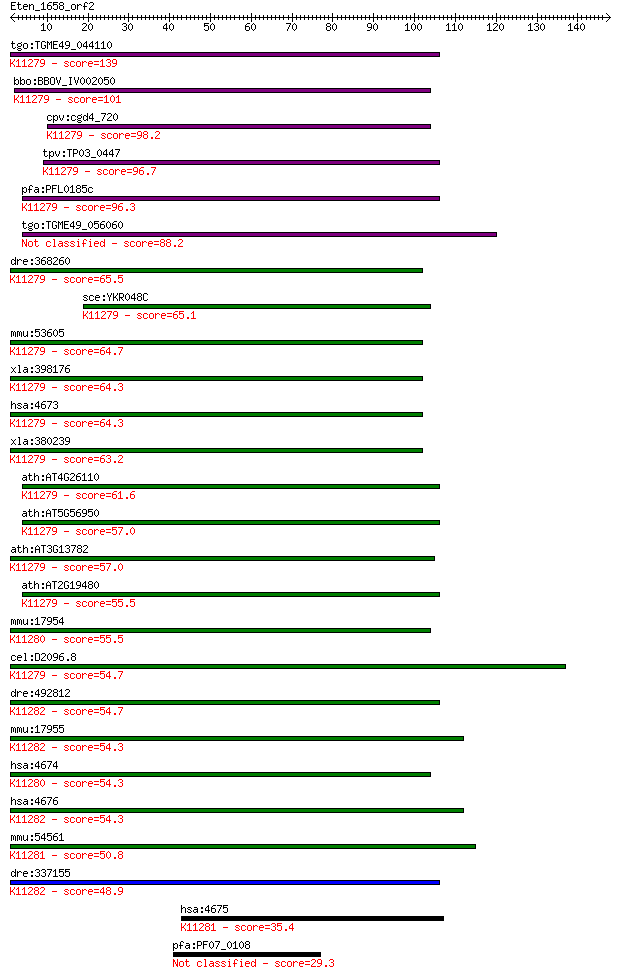
tgo:TGME49_044110 nucleosome assembly protein, putative ; K112... 139 3e-33
bbo:BBOV_IV002050 21.m02796; nucleosome assembly protein 1; K1... 101 9e-22
cpv:cgd4_720 nucleosome assembly protein 19075338; besthit Py ... 98.2 8e-21
tpv:TP03_0447 nucleosome assembly protein 1; K11279 nucleosome... 96.7 2e-20
pfa:PFL0185c nucleosome assembly protein 1, putative; K11279 n... 96.3 3e-20
tgo:TGME49_056060 nucleosome assembly protein, putative (EC:3.... 88.2 9e-18
dre:368260 nap1l1, cb716, sb:cb13, wu:fb83g07; nucleosome asse... 65.5 7e-11
sce:YKR048C NAP1; Nap1p; K11279 nucleosome assembly protein 1-... 65.1 8e-11
mmu:53605 Nap1l1, AA407126, AI256722, D10Ertd68e, NAP-1; nucle... 64.7 1e-10
xla:398176 nap1l1-a, MGC81048, NAP-1, nap1, nap1-A, xNAP-1, xN... 64.3 1e-10
hsa:4673 NAP1L1, FLJ16112, MGC23410, MGC8688, NAP1, NAP1L, NRP... 64.3 1e-10
xla:380239 nap1l1-b, MGC53871, NAP1, nap1-B, nap1l1, xNAP-1, x... 63.2 3e-10
ath:AT4G26110 NAP1;1; NAP1;1 (NUCLEOSOME ASSEMBLY PROTEIN1;1);... 61.6 9e-10
ath:AT5G56950 NAP1;3; NAP1;3 (NUCLEOSOME ASSEMBLY PROTEIN 1;3)... 57.0 2e-08
ath:AT3G13782 NAP1;4; NAP1;4 (NUCLEOSOME ASSEMBLY PROTEIN1;4);... 57.0 2e-08
ath:AT2G19480 NAP1;2; NAP1;2 (NUCLEOSOME ASSEMBLY PROTEIN 1;2)... 55.5 6e-08
mmu:17954 Nap1l2, Bpx; nucleosome assembly protein 1-like 2; K... 55.5 6e-08
cel:D2096.8 hypothetical protein; K11279 nucleosome assembly p... 54.7 9e-08
dre:492812 nap1l4b, zgc:101613; nucleosome assembly protein 1-... 54.7 1e-07
mmu:17955 Nap1l4, 2810410H14Rik, AI316776, D7Wsu30e, Nap2; nuc... 54.3 1e-07
hsa:4674 NAP1L2, BPX, MGC26243; nucleosome assembly protein 1-... 54.3 1e-07
hsa:4676 NAP1L4, MGC4565, NAP1L4b, NAP2, NAP2L, hNAP2; nucleos... 54.3 1e-07
mmu:54561 Nap1l3, MB20; nucleosome assembly protein 1-like 3; ... 50.8 1e-06
dre:337155 nap1l4a, cb910, nap1l4, wu:fa99c07; nucleosome asse... 48.9 5e-06
hsa:4675 NAP1L3, MB20, MGC26312, NPL3; nucleosome assembly pro... 35.4 0.064
pfa:PF07_0108 conserved Plasmodium protein, unknown function 29.3 5.0
> tgo:TGME49_044110 nucleosome assembly protein, putative ; K11279
nucleosome assembly protein 1-like 1
Length=433
Score = 139 bits (350), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 63/105 (60%), Positives = 82/105 (78%), Gaps = 0/105 (0%)
Query 1 GAELLRSSSSKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPP 60
GAEL + S+ I W ++VTKKV+ RKQRN+KT++TRTV E V+ SFFN F DHEIP
Sbjct 276 GAELTSTKSTPIAWKEGKDVTKKVVTRKQRNRKTRQTRTVEEVVDNESFFNFFTDHEIPS 335
Query 61 DHKLELMDEKEAEELQLVVEADYDIGIRIRDKIIPRAVDWFLGQV 105
+ K+E M +K+ ELQ++VEADYDIG+ IRDKIIP+AV+WFLG+
Sbjct 336 EEKIETMSDKQIGELQMIVEADYDIGVTIRDKIIPQAVEWFLGEA 380
> bbo:BBOV_IV002050 21.m02796; nucleosome assembly protein 1;
K11279 nucleosome assembly protein 1-like 1
Length=297
Score = 101 bits (251), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 47/102 (46%), Positives = 70/102 (68%), Gaps = 0/102 (0%)
Query 2 AELLRSSSSKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPPD 61
A L + +++I W P+++VT++ + + QR+K+TKETRT TE + SFF F E+P +
Sbjct 164 ALLQGTEATEINWYPDKDVTRQTVTKIQRHKRTKETRTKTEVEDQPSFFRFFTTQEVPSN 223
Query 62 HKLELMDEKEAEELQLVVEADYDIGIRIRDKIIPRAVDWFLG 103
L M ++E EL++ VE DYDIGI IRDK+IP A+ W+LG
Sbjct 224 EALSRMTKQEIAELEMYVEEDYDIGIVIRDKLIPEAIYWYLG 265
> cpv:cgd4_720 nucleosome assembly protein 19075338; besthit Py
23481872 ; K11279 nucleosome assembly protein 1-like 1
Length=347
Score = 98.2 bits (243), Expect = 8e-21, Method: Compositional matrix adjust.
Identities = 47/94 (50%), Positives = 65/94 (69%), Gaps = 0/94 (0%)
Query 10 SKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPPDHKLELMDE 69
SKIEW R+ T++V++R+QR+K++KE R VTETV SFFN F +P D +L MD
Sbjct 181 SKIEWKQGRDPTQEVVVRRQRHKQSKEVRIVTETVHRESFFNFFKSLNVPSDEELAKMDR 240
Query 70 KEAEELQLVVEADYDIGIRIRDKIIPRAVDWFLG 103
+ EL+ VE DY++GI IRDK+IP ++ WF G
Sbjct 241 FDIMELESTVETDYEMGIFIRDKLIPYSLYWFTG 274
> tpv:TP03_0447 nucleosome assembly protein 1; K11279 nucleosome
assembly protein 1-like 1
Length=292
Score = 96.7 bits (239), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 44/97 (45%), Positives = 66/97 (68%), Gaps = 0/97 (0%)
Query 9 SSKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPPDHKLELMD 68
+++I W P+++VTK+ + + QR+KKTKETRT + + SFF F E+P +L M
Sbjct 172 ATEINWLPDKDVTKQTVTKIQRHKKTKETRTRVDLEDKPSFFRFFTGQEVPTTEELNKMS 231
Query 69 EKEAEELQLVVEADYDIGIRIRDKIIPRAVDWFLGQV 105
+ E EL++ VE DYDIGI +RDK++P A+ W+LG V
Sbjct 232 KLEISELEMYVEEDYDIGIVLRDKLVPEAIYWYLGVV 268
> pfa:PFL0185c nucleosome assembly protein 1, putative; K11279
nucleosome assembly protein 1-like 1
Length=347
Score = 96.3 bits (238), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 36/102 (35%), Positives = 72/102 (70%), Gaps = 0/102 (0%)
Query 4 LLRSSSSKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPPDHK 63
LL + ++ I+W +N+ KK +++KQ NK ++E +TV +TV +SFF+ F H++P +
Sbjct 180 LLHTEATVIDWYDNKNILKKNVVKKQHNKNSREVKTVQQTVNRDSFFHFFTSHKVPNSNV 239
Query 64 LELMDEKEAEELQLVVEADYDIGIRIRDKIIPRAVDWFLGQV 105
++ + + E +L++++E DY++ + I+++IIP AVD++LG +
Sbjct 240 IKQLSKHEVAQLEMIIEGDYEVALTIKERIIPYAVDYYLGII 281
> tgo:TGME49_056060 nucleosome assembly protein, putative (EC:3.4.21.72)
Length=812
Score = 88.2 bits (217), Expect = 9e-18, Method: Compositional matrix adjust.
Identities = 46/116 (39%), Positives = 68/116 (58%), Gaps = 3/116 (2%)
Query 4 LLRSSSSKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPPDHK 63
+ R+ + I W +VTK ++RKQRN +T RTV E SFF+ F D +P D +
Sbjct 668 VSRTVGTSIVWRENMDVTKSAVVRKQRNVRTNNVRTVHEEKHEKSFFSFFCDSVVPEDDE 727
Query 64 LELMDEKEAEELQLVVEADYDIGIRIRDKIIPRAVDWFLGQVDDDSDFEDYDTNEE 119
LE M+E+E E+ ++ +YD G+ +RD IIP A++W+LG + YD NEE
Sbjct 728 LEAMNEEEQSEIFEKLKLEYDAGVTLRDLIIPYALEWYLGSA---VETLGYDDNEE 780
> dre:368260 nap1l1, cb716, sb:cb13, wu:fb83g07; nucleosome assembly
protein 1, like 1; K11279 nucleosome assembly protein
1-like 1
Length=385
Score = 65.5 bits (158), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 40/101 (39%), Positives = 62/101 (61%), Gaps = 7/101 (6%)
Query 1 GAELLRSSSSKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPP 60
G E++ + I+W +NVT K I +KQ++K RTVT+TV +SFFN F E+P
Sbjct 241 GPEIMGCTGCTIDWTKGKNVTLKTIKKKQKHKGRGTVRTVTKTVPNDSFFNFFSPPEVPE 300
Query 61 DHKLELMDEKEAEELQLVVEADYDIGIRIRDKIIPRAVDWF 101
+L+ E+ + V+ AD++IG IR++I+PRAV +F
Sbjct 301 SGELD-------EDSEAVLAADFEIGHFIRERIVPRAVLYF 334
> sce:YKR048C NAP1; Nap1p; K11279 nucleosome assembly protein
1-like 1
Length=417
Score = 65.1 bits (157), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 40/85 (47%), Positives = 54/85 (63%), Gaps = 4/85 (4%)
Query 19 NVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPPDHKLELMDEKEAEELQLV 78
NVT + +RKQRNK TK+ RT+ + SFFN F PP + E DE+ E+L+
Sbjct 282 NVTVDLEMRKQRNKTTKQVRTIEKITPIESFFNFFD----PPKIQNEDQDEELEEDLEER 337
Query 79 VEADYDIGIRIRDKIIPRAVDWFLG 103
+ DY IG +++DK+IPRAVDWF G
Sbjct 338 LALDYSIGEQLKDKLIPRAVDWFTG 362
> mmu:53605 Nap1l1, AA407126, AI256722, D10Ertd68e, NAP-1; nucleosome
assembly protein 1-like 1; K11279 nucleosome assembly
protein 1-like 1
Length=418
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 40/101 (39%), Positives = 64/101 (63%), Gaps = 7/101 (6%)
Query 1 GAELLRSSSSKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPP 60
G E++ + +I+W +NVT K I +KQ++K RTVT+TV +SFFN F E+P
Sbjct 276 GPEIMGCTGCQIDWKKGKNVTLKTIKKKQKHKGRGTVRTVTKTVSNDSFFNFFAPPEVPE 335
Query 61 DHKLELMDEKEAEELQLVVEADYDIGIRIRDKIIPRAVDWF 101
+ L+ D+ EA ++ AD++IG +R++IIPR+V +F
Sbjct 336 NGDLD--DDAEA-----ILAADFEIGHFLRERIIPRSVLYF 369
> xla:398176 nap1l1-a, MGC81048, NAP-1, nap1, nap1-A, xNAP-1,
xNAP1, xNAP1L-A, xnap1l, xnap1l1; nucleosome assembly protein
1-like 1; K11279 nucleosome assembly protein 1-like 1
Length=304
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 40/101 (39%), Positives = 64/101 (63%), Gaps = 7/101 (6%)
Query 1 GAELLRSSSSKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPP 60
G E++ + I+W +NVT K I +KQ++K RTVT+TV +SFFN F E+P
Sbjct 161 GPEIMGCTGCLIDWKKGKNVTLKTIKKKQKHKGRGTVRTVTKTVPNDSFFNFFTPPEVPE 220
Query 61 DHKLELMDEKEAEELQLVVEADYDIGIRIRDKIIPRAVDWF 101
+ +L+ D+ EA ++ AD++IG +R++IIPR+V +F
Sbjct 221 NGELD--DDAEA-----ILTADFEIGHFLRERIIPRSVLYF 254
> hsa:4673 NAP1L1, FLJ16112, MGC23410, MGC8688, NAP1, NAP1L, NRP;
nucleosome assembly protein 1-like 1; K11279 nucleosome
assembly protein 1-like 1
Length=391
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 40/101 (39%), Positives = 63/101 (62%), Gaps = 7/101 (6%)
Query 1 GAELLRSSSSKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPP 60
G E++ + +I+W +NVT K I +KQ++K RTVT+TV +SFFN F E+P
Sbjct 249 GPEIMGCTGCQIDWKKGKNVTLKTIKKKQKHKGRGTVRTVTKTVSNDSFFNFFAPPEVPE 308
Query 61 DHKLELMDEKEAEELQLVVEADYDIGIRIRDKIIPRAVDWF 101
L+ D+ EA ++ AD++IG +R++IIPR+V +F
Sbjct 309 SGDLD--DDAEA-----ILAADFEIGHFLRERIIPRSVLYF 342
> xla:380239 nap1l1-b, MGC53871, NAP1, nap1-B, nap1l1, xNAP-1,
xNAP1L-B, xnap1l, xnap1l1; nucleosome assembly protein 1-like
1; K11279 nucleosome assembly protein 1-like 1
Length=393
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 40/101 (39%), Positives = 64/101 (63%), Gaps = 7/101 (6%)
Query 1 GAELLRSSSSKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPP 60
G E++ + I+W +NVT K I +KQ++K RTVT+TV +SFFN F E+P
Sbjct 250 GPEIMGCTGCLIDWKKGKNVTLKTIKKKQKHKGRGTVRTVTKTVPNDSFFNFFSPPEVPE 309
Query 61 DHKLELMDEKEAEELQLVVEADYDIGIRIRDKIIPRAVDWF 101
+ +L+ D+ EA ++ AD++IG +R++IIPR+V +F
Sbjct 310 NGELD--DDAEA-----ILTADFEIGHFLRERIIPRSVLYF 343
> ath:AT4G26110 NAP1;1; NAP1;1 (NUCLEOSOME ASSEMBLY PROTEIN1;1);
DNA binding; K11279 nucleosome assembly protein 1-like 1
Length=359
Score = 61.6 bits (148), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 43/102 (42%), Positives = 69/102 (67%), Gaps = 3/102 (2%)
Query 4 LLRSSSSKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPPDHK 63
L ++ ++I+W P + +T+K++ +K+ K +K T+ +T+ + SFFN F E+P +
Sbjct 201 LEKAMGTEIDWYPGKCLTQKIL-KKKPKKGSKNTKPITKLEDCESFFNFFSPPEVP--DE 257
Query 64 LELMDEKEAEELQLVVEADYDIGIRIRDKIIPRAVDWFLGQV 105
E +DE+ AE+LQ ++E DYDIG IR+KIIPRAV WF G+
Sbjct 258 DEDIDEERAEDLQNLMEQDYDIGSTIREKIIPRAVSWFTGEA 299
> ath:AT5G56950 NAP1;3; NAP1;3 (NUCLEOSOME ASSEMBLY PROTEIN 1;3);
DNA binding; K11279 nucleosome assembly protein 1-like 1
Length=374
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 42/102 (41%), Positives = 66/102 (64%), Gaps = 3/102 (2%)
Query 4 LLRSSSSKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPPDHK 63
L ++ ++I+W P + +T+K++ +K+ K K + +T+T + SFFN F ++P
Sbjct 200 LEKAIGTEIDWYPGKCLTQKIL-KKKPKKGAKNAKPITKTEDCESFFNFFNPPQVP--DD 256
Query 64 LELMDEKEAEELQLVVEADYDIGIRIRDKIIPRAVDWFLGQV 105
E +DE+ AEELQ ++E DYDIG IR+KIIP AV WF G+
Sbjct 257 DEDIDEERAEELQNLMEQDYDIGSTIREKIIPHAVSWFTGEA 298
> ath:AT3G13782 NAP1;4; NAP1;4 (NUCLEOSOME ASSEMBLY PROTEIN1;4);
DNA binding / chromatin binding; K11279 nucleosome assembly
protein 1-like 1
Length=329
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 42/109 (38%), Positives = 64/109 (58%), Gaps = 5/109 (4%)
Query 1 GAELLRSSSSKIEWAPERNVTKKVIIRKQRNKKTKETRTV--TETVETNSFFNLFVDHEI 58
G L + + IEW P + +T KV+++K+ K K+ + T+T SFFN F EI
Sbjct 200 GPVLEKVIGTDIEWFPGKCLTHKVVVKKKTKKGPKKVNNIPMTKTENCESFFNFFKPPEI 259
Query 59 PPDHKLELMDEKEA---EELQLVVEADYDIGIRIRDKIIPRAVDWFLGQ 104
P +++ D+ + EELQ +++ DYDI + IRDK+IP AV WF G+
Sbjct 260 PEIDEVDDYDDFDTIMTEELQNLMDQDYDIAVTIRDKLIPHAVSWFTGE 308
> ath:AT2G19480 NAP1;2; NAP1;2 (NUCLEOSOME ASSEMBLY PROTEIN 1;2);
DNA binding; K11279 nucleosome assembly protein 1-like 1
Length=373
Score = 55.5 bits (132), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 37/102 (36%), Positives = 62/102 (60%), Gaps = 3/102 (2%)
Query 4 LLRSSSSKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPPDHK 63
L ++ ++IEW P + +T+K++ +K+ K +K T+ +T+T + SFFN F ++P D +
Sbjct 200 LEKALGTEIEWYPGKCLTQKIL-KKKPKKGSKNTKPITKTEDCESFFNFFSPPQVPDDDE 258
Query 64 LELMDEKEAEELQLVVEADYDIGIRIRDKIIPRAVDWFLGQV 105
D + + Q+ E DYDIG I++KII AV WF G+
Sbjct 259 DLDDDMADELQGQM--EHDYDIGSTIKEKIISHAVSWFTGEA 298
> mmu:17954 Nap1l2, Bpx; nucleosome assembly protein 1-like 2;
K11280 nucleosome assembly protein 1-like 2
Length=460
Score = 55.5 bits (132), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 35/103 (33%), Positives = 51/103 (49%), Gaps = 14/103 (13%)
Query 1 GAELLRSSSSKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPP 60
G + ++ I+W +NVT + I +KQR++ RTVTE +SFFN F H I
Sbjct 322 GTAIEYATGCDIDWNEGKNVTLRTIKKKQRHRVWGTVRTVTEDFPKDSFFNFFSPHGI-- 379
Query 61 DHKLELMDEKEAEELQLVVEADYDIGIRIRDKIIPRAVDWFLG 103
L DE + D+ +G +R IIPR+V +F G
Sbjct 380 --SLNGGDEND----------DFLLGHNLRTYIIPRSVLFFSG 410
> cel:D2096.8 hypothetical protein; K11279 nucleosome assembly
protein 1-like 1
Length=316
Score = 54.7 bits (130), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 44/136 (32%), Positives = 74/136 (54%), Gaps = 13/136 (9%)
Query 1 GAELLRSSSSKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPP 60
G ++R+ IEW + K ++K++ K + +T+TV+ +SFFN F PP
Sbjct 193 GPHVIRAVGDTIEWE-DGKNVTKKAVKKKQKKGANAGKFLTKTVKADSFFNFFE----PP 247
Query 61 DHKLELMDEKEAEELQLVVEADYDIGIRIRDKIIPRAVDWFLGQVDDDSDFEDYDTNEEE 120
K E ++++ E+ + +E DY++G IRD IIPRAV ++ G++ D F +
Sbjct 248 KSKDERNEDEDDEQAEEFLELDYEMGQAIRDTIIPRAVLFYTGELQSDDMF--------D 299
Query 121 FDTEDSDEEDDSSDED 136
F ED D+ D SD++
Sbjct 300 FPGEDGDDVSDFSDDE 315
> dre:492812 nap1l4b, zgc:101613; nucleosome assembly protein
1-like 4b; K11282 nucleosome assembly protein 1-like 4
Length=367
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 34/105 (32%), Positives = 60/105 (57%), Gaps = 7/105 (6%)
Query 1 GAELLRSSSSKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPP 60
G E++ +I+W ++VT K I +KQ++K R +T+ V +SFFN F ++ P
Sbjct 223 GPEIIDCEGCEIDWQKGKDVTVKTIKKKQKHKGRGTARVITKQVPNDSFFNFFNPIKVSP 282
Query 61 DHKLELMDEKEAEELQLVVEADYDIGIRIRDKIIPRAVDWFLGQV 105
D +L+ E+ + + D++IG R++I+PRAV +F G+
Sbjct 283 DKELD-------EDSEYTLATDFEIGHFFRERIVPRAVLYFTGEA 320
> mmu:17955 Nap1l4, 2810410H14Rik, AI316776, D7Wsu30e, Nap2; nucleosome
assembly protein 1-like 4; K11282 nucleosome assembly
protein 1-like 4
Length=375
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 39/112 (34%), Positives = 64/112 (57%), Gaps = 7/112 (6%)
Query 1 GAELLRSSSSKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPP 60
G E++ I+W +NVT K I +KQ++K RT+T+ V SFFN F +
Sbjct 241 GPEIVDCDGCTIDWKKGKNVTVKTIKKKQKHKGRGTVRTITKQVPNESFFNFFSPLKASG 300
Query 61 DHKLELMDEKEAEELQLVVEADYDIGIRIRDKIIPRAVDWFLGQ-VDDDSDF 111
D E +DE + + + +D++IG R++I+PRAV +F G+ ++DD +F
Sbjct 301 DG--ESLDE----DSEFTLASDFEIGHFFRERIVPRAVLYFTGEAIEDDDNF 346
> hsa:4674 NAP1L2, BPX, MGC26243; nucleosome assembly protein
1-like 2; K11280 nucleosome assembly protein 1-like 2
Length=460
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 34/103 (33%), Positives = 52/103 (50%), Gaps = 14/103 (13%)
Query 1 GAELLRSSSSKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPP 60
G + S+ +I+W +NVT K I +KQ+++ RTVTE +SFFN F H I
Sbjct 322 GTAIEYSTGCEIDWNEGKNVTLKTIKKKQKHRIWGTIRTVTEDFPKDSFFNFFSPHGITS 381
Query 61 DHKLELMDEKEAEELQLVVEADYDIGIRIRDKIIPRAVDWFLG 103
+ + D + D+ +G +R IIPR+V +F G
Sbjct 382 NGR----DGND----------DFLLGHNLRTYIIPRSVLFFSG 410
> hsa:4676 NAP1L4, MGC4565, NAP1L4b, NAP2, NAP2L, hNAP2; nucleosome
assembly protein 1-like 4; K11282 nucleosome assembly
protein 1-like 4
Length=375
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 38/114 (33%), Positives = 65/114 (57%), Gaps = 11/114 (9%)
Query 1 GAELLRSSSSKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPP 60
G E++ I+W +NVT K I +KQ++K RT+T+ V SFFN F
Sbjct 241 GPEIVDCDGCTIDWKKGKNVTVKTIKKKQKHKGRGTVRTITKQVPNESFFNFF------- 293
Query 61 DHKLELMDEKEA--EELQLVVEADYDIGIRIRDKIIPRAVDWFLGQ-VDDDSDF 111
+ L+ + E+ E+ + + +D++IG R++I+PRAV +F G+ ++DD +F
Sbjct 294 -NPLKASGDGESLDEDSEFTLASDFEIGHFFRERIVPRAVLYFTGEAIEDDDNF 346
> mmu:54561 Nap1l3, MB20; nucleosome assembly protein 1-like 3;
K11281 nucleosome assembly protein 1-like 3
Length=544
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 35/116 (30%), Positives = 56/116 (48%), Gaps = 11/116 (9%)
Query 1 GAELLRSSSSKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPP 60
G E+ KI+W ++VT V + R T E V SFFN F EIP
Sbjct 431 GWEIEECKGCKIDWRRGKDVT--VTTTRSRPGITGEIEVQPRVVPNASFFNFFSPPEIPL 488
Query 61 DHKLELMDEKEAEELQLVVEADYDIGIRIRDKIIPRAVDWFLGQVDDD--SDFEDY 114
KLE ++ +++ D++IG + D +I +++ +F G+++D DF DY
Sbjct 489 IGKLEPRED-------AILDEDFEIGQILHDNVILKSIYYFTGEINDPYYHDFRDY 537
> dre:337155 nap1l4a, cb910, nap1l4, wu:fa99c07; nucleosome assembly
protein 1-like 4a; K11282 nucleosome assembly protein
1-like 4
Length=349
Score = 48.9 bits (115), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 36/105 (34%), Positives = 60/105 (57%), Gaps = 8/105 (7%)
Query 1 GAELLRSSSSKIEWAPERNVTKKVIIRKQRNKKTKETRTVTETVETNSFFNLFVDHEIPP 60
G E++ KI+W ++VT K++ +KQ++K RTV++ + +SFFN F + P
Sbjct 217 GPEIVDCEGCKIDWHKGKDVTVKIVKKKQKHKGRGTIRTVSKEIPQDSFFNFFNPVKASP 276
Query 61 DHKLELMDEKEAEELQLVVEADYDIGIRIRDKIIPRAVDWFLGQV 105
D MD E+L + D++IG R+++IPRAV +F G+
Sbjct 277 DG----MD----EDLDFTLATDFEIGHFFRERVIPRAVLYFTGEA 313
> hsa:4675 NAP1L3, MB20, MGC26312, NPL3; nucleosome assembly protein
1-like 3; K11281 nucleosome assembly protein 1-like 3
Length=506
Score = 35.4 bits (80), Expect = 0.064, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 35/64 (54%), Gaps = 7/64 (10%)
Query 43 TVETNSFFNLFVDHEIPPDHKLELMDEKEAEELQLVVEADYDIGIRIRDKIIPRAVDWFL 102
V SFFN F EIP KLE ++ +++ D++IG + D +I +++ ++
Sbjct 433 VVPNASFFNFFSPPEIPMIGKLEPRED-------AILDEDFEIGQILHDNVILKSIYYYT 485
Query 103 GQVD 106
G+V+
Sbjct 486 GEVN 489
> pfa:PF07_0108 conserved Plasmodium protein, unknown function
Length=688
Score = 29.3 bits (64), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 26/36 (72%), Gaps = 0/36 (0%)
Query 41 TETVETNSFFNLFVDHEIPPDHKLELMDEKEAEELQ 76
T+ V+TN++FN FV+H+ + + EL+ +K+ +E Q
Sbjct 137 TDNVQTNNYFNEFVNHQENKETEGELICDKKKKEEQ 172
Lambda K H
0.311 0.132 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2938175820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40