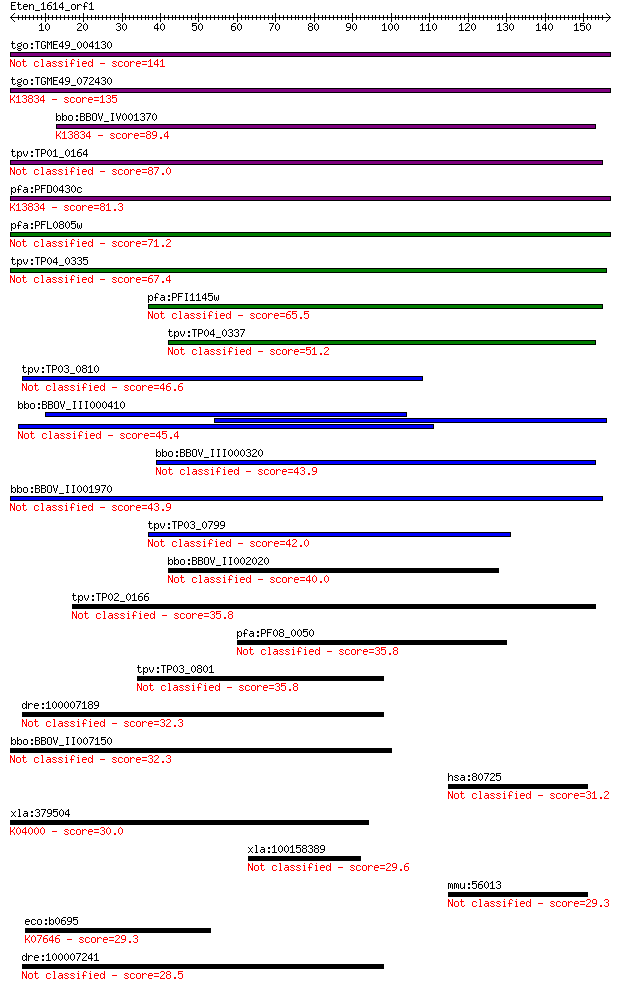
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1614_orf1
Length=156
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_004130 membrane-attack complex / perforin domain-co... 141 7e-34
tgo:TGME49_072430 membrane-attack complex / perforin domain-co... 135 5e-32
bbo:BBOV_IV001370 21.m02755; MAC/perforin domain containing pr... 89.4 5e-18
tpv:TP01_0164 hypothetical protein 87.0 2e-17
pfa:PFD0430c MAC/Perforin, putative; K13834 sporozoite microne... 81.3 1e-15
pfa:PFL0805w MAC/Perforin, putative 71.2 1e-12
tpv:TP04_0335 hypothetical protein 67.4 2e-11
pfa:PFI1145w MAC/Perforin, putative 65.5 7e-11
tpv:TP04_0337 hypothetical protein 51.2 1e-06
tpv:TP03_0810 hypothetical protein 46.6 3e-05
bbo:BBOV_III000410 hypothetical protein 45.4 7e-05
bbo:BBOV_III000320 17.m10445; hypothetical protein 43.9 2e-04
bbo:BBOV_II001970 18.m09950; mac/perforin domain containing me... 43.9 2e-04
tpv:TP03_0799 hypothetical protein 42.0 7e-04
bbo:BBOV_II002020 18.m06160; mac/perforin domain containing pr... 40.0 0.003
tpv:TP02_0166 hypothetical protein 35.8 0.052
pfa:PF08_0050 MAC/Perforin, putative 35.8 0.052
tpv:TP03_0801 hypothetical protein 35.8 0.066
dre:100007189 Perforin-1-like 32.3 0.62
bbo:BBOV_II007150 18.m06592; mac/perforin domain containing pr... 32.3 0.67
hsa:80725 SRCIN1, KIAA1684, P140, SNIP; SRC kinase signaling i... 31.2 1.3
xla:379504 c9, MGC64276; complement component 9; K04000 comple... 30.0 3.3
xla:100158389 hypothetical protein LOC100158389 29.6 4.6
mmu:56013 Srcin1, P140, SNIP, mKIAA1684, p140Cap; SRC kinase s... 29.3 4.8
eco:b0695 kdpD, ECK0683, JW0683, kac; fused sensory histidine ... 29.3 5.2
dre:100007241 Perforin-1-like 28.5 9.9
> tgo:TGME49_004130 membrane-attack complex / perforin domain-containing
protein
Length=1054
Score = 141 bits (356), Expect = 7e-34, Method: Compositional matrix adjust.
Identities = 73/156 (46%), Positives = 105/156 (67%), Gaps = 0/156 (0%)
Query 1 NVKSALKATFGVVSGGVSSNVSKEAEQREQLKHFQYETETLIVGGRVPKDVSDPDSMAEW 60
+VK+ LK G VSGG S + Q + ETL++GGR P +VSDP ++A W
Sbjct 599 DVKTQLKMQLGGVSGGAGQGTSSKKNQSSSEYQMNVQKETLVIGGRPPGNVSDPAALAAW 658
Query 61 SDTVEELPMPVKITIQSLSYLLPEEKKEAFEKASRFYAEAVGMSRADLNAMGGKVESIGE 120
+DTVEELPMPVK +Q L +LLP EK+EAF++A FY+++VG++ DL+A+ G ++ +
Sbjct 659 ADTVEELPMPVKFEVQPLYHLLPVEKQEAFKQAVTFYSKSVGLTPQDLSALTGVTRNLPK 718
Query 121 VLRKATQVTYAGDPPGYAFCAPHERVLLGFALQLNF 156
L +ATQV ++G PPG+A C + V+LGFA+ LNF
Sbjct 719 QLTQATQVAWSGPPPGFAKCPEGQVVILGFAMHLNF 754
> tgo:TGME49_072430 membrane-attack complex / perforin domain-containing
protein ; K13834 sporozoite microneme protein 2
Length=854
Score = 135 bits (340), Expect = 5e-32, Method: Compositional matrix adjust.
Identities = 69/156 (44%), Positives = 100/156 (64%), Gaps = 0/156 (0%)
Query 1 NVKSALKATFGVVSGGVSSNVSKEAEQREQLKHFQYETETLIVGGRVPKDVSDPDSMAEW 60
+VK+ +K G S G SS V + QL+ E E L++GG+ P DVSDP ++A W
Sbjct 462 DVKAEVKLMLGGFSAGASSQVKTNQDSASQLRSLNVEKEALVIGGKPPADVSDPKAIAAW 521
Query 61 SDTVEELPMPVKITIQSLSYLLPEEKKEAFEKASRFYAEAVGMSRADLNAMGGKVESIGE 120
+++V+ LPMPVK+ + L LLPE+K+EAF A +Y++A GMS D+ ++ G SI +
Sbjct 522 ANSVDALPMPVKLELLPLQNLLPEDKREAFTHAVTYYSKAFGMSAMDVQSLEGTARSIQD 581
Query 121 VLRKATQVTYAGDPPGYAFCAPHERVLLGFALQLNF 156
VL+ TQ+ +AG PPGYA C + VL GFA++ NF
Sbjct 582 VLKDVTQIAWAGAPPGYARCPREQVVLFGFAMRFNF 617
> bbo:BBOV_IV001370 21.m02755; MAC/perforin domain containing
protein; K13834 sporozoite microneme protein 2
Length=978
Score = 89.4 bits (220), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 48/141 (34%), Positives = 78/141 (55%), Gaps = 3/141 (2%)
Query 13 VSGGVSSNVSKEAEQREQLKHFQYETETLIVGGRVPKDVSDPDSMAEWSDTVEELPMPVK 72
V GG S K+ E Q E +++GG VP D +D +M EW+ ++ PMP+K
Sbjct 563 VGGGFSH--EKKEESSSNFSSLQTEKLVIVIGGDVPTDGTDKTTMLEWTKSLYRKPMPIK 620
Query 73 ITIQSLSYLLP-EEKKEAFEKASRFYAEAVGMSRADLNAMGGKVESIGEVLRKATQVTYA 131
+ I+S+ L+ EKK++F+ A ++Y+EA G+S +L G+ I + R T VTY
Sbjct 621 VNIESIKTLIDGHEKKKSFDTALKYYSEAYGISPEELYRREGRKMGIAGLARHGTPVTYY 680
Query 132 GDPPGYAFCAPHERVLLGFAL 152
G G A C + +++GF++
Sbjct 681 GTTGGSAVCPNGKVIMMGFSV 701
> tpv:TP01_0164 hypothetical protein
Length=1182
Score = 87.0 bits (214), Expect = 2e-17, Method: Composition-based stats.
Identities = 49/159 (30%), Positives = 88/159 (55%), Gaps = 8/159 (5%)
Query 1 NVKSALKATFGVV-----SGGVSSNVSKEAEQREQLKHFQYETETLIVGGRVPKDVSDPD 55
NV +A+KA+ V GG +S + E Q + +Y+ + L++GG D + +
Sbjct 807 NVDAAVKASISPVMVDSLQGGFAS--TSEKASLSQSNNLKYDKQVLVIGGDGLVDSKNAN 864
Query 56 SMAEWSDTVEELPMPVKITIQSLSYLLPEEKKEAFEKASRFYAEAVGMSRADLNAMGGKV 115
S+ W+ + + PMP+KI ++S+ LL +K+E F++A +FY+E G+S ++ GK
Sbjct 865 SLNNWAKELYKRPMPIKIKLESIKSLL-GKKRELFDEALKFYSETYGVSPDEIYKKFGKE 923
Query 116 ESIGEVLRKATQVTYAGDPPGYAFCAPHERVLLGFALQL 154
I V+ K Q+ Y+G+ G A C +++GF+L +
Sbjct 924 FGIASVIEKGHQIVYSGNKSGSAICPNKTVIIMGFSLSI 962
> pfa:PFD0430c MAC/Perforin, putative; K13834 sporozoite microneme
protein 2
Length=842
Score = 81.3 bits (199), Expect = 1e-15, Method: Composition-based stats.
Identities = 48/157 (30%), Positives = 83/157 (52%), Gaps = 3/157 (1%)
Query 1 NVKSALKATFGVVSGGVSSNVSKEAEQREQLKHFQYETETLIVGGRVPKDVSDPDSMAEW 60
+VK+ ++A FG S G S++V+ + + + +++GG KDV+ +++ EW
Sbjct 468 SVKAKIQAQFGFGSAGGSTDVNSSNSSANDEQSYDMNEQLIVIGGNPIKDVTKEENLFEW 527
Query 61 SDTVEELPMPVKITIQSLS-YLLPEEKKEAFEKASRFYAEAVGMSRADLNAMGGKVESIG 119
S TV PMP+ I + +S ++ KE+++KA +Y+ G+S D M + I
Sbjct 528 SKTVTNHPMPINIKLTPISDSFDSDDLKESYDKAIIYYSRLYGLSPHD--TMQKDDKDII 585
Query 120 EVLRKATQVTYAGDPPGYAFCAPHERVLLGFALQLNF 156
++L A VT PP A C + V+ GF+L+ NF
Sbjct 586 KILTNADTVTKNSAPPINAQCPHGKVVMFGFSLKQNF 622
> pfa:PFL0805w MAC/Perforin, putative
Length=1073
Score = 71.2 bits (173), Expect = 1e-12, Method: Composition-based stats.
Identities = 45/157 (28%), Positives = 74/157 (47%), Gaps = 5/157 (3%)
Query 1 NVKSALKATFGVVSGGVSSNVSKEAEQREQLKHFQYETETLIVGGRVPKDVSDPDSMAEW 60
NVK+ F + G+SS KEA++ L F+ I+GG +V++ +W
Sbjct 693 NVKTFFNIYFHKM--GLSSAFQKEAQK--ILNKFRISKHIAILGGNPGLNVNNTSFFEKW 748
Query 61 SDTVEELPMPVKITIQSLSYLLPEEKK-EAFEKASRFYAEAVGMSRADLNAMGGKVESIG 119
++ MP++ + S+ + + +A++ A FY G+ + S+G
Sbjct 749 VHSINTNSMPIRTKLLPFSFFMDDNDMIQAYKDALIFYGLTYGLQLFGQEKYNHNIISVG 808
Query 120 EVLRKATQVTYAGDPPGYAFCAPHERVLLGFALQLNF 156
E L K TQ YAG PPG C +L+GF++ LNF
Sbjct 809 EYLEKCTQKLYAGPPPGLLTCPIGTTILMGFSINLNF 845
> tpv:TP04_0335 hypothetical protein
Length=441
Score = 67.4 bits (163), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 44/157 (28%), Positives = 73/157 (46%), Gaps = 2/157 (1%)
Query 1 NVKSALKATFGVVSGGVSSNVSKEAEQREQLKHFQYETETLIVGGRVPKDVSDPDSMAEW 60
+V +A+ A VS ++ VSK E + + + ++GG P P S+ +W
Sbjct 271 DVNAAIGAVISGVSANIAVGVSKSEESEIKQLKKSSKLKFSVLGGIHPDRNISPTSIRKW 330
Query 61 SDTVEELPMPVKITIQSLSYLLPEEKKEAFEKASRFYAEAVGMSRADLNAMGGKVESIGE 120
TV PMP+KI ++S+S LP +F++A FY D+ ++ ++
Sbjct 331 KSTVPRYPMPIKIDVESISTFLPRSYYNSFKEALSFYITLNKALPWDIEQKNKRLMTMKS 390
Query 121 VLRKATQ--VTYAGDPPGYAFCAPHERVLLGFALQLN 155
+L + Q VT D A C +RVL GF + +N
Sbjct 391 LLVNSKQILVTNKNDELPVAKCQDGKRVLFGFIININ 427
> pfa:PFI1145w MAC/Perforin, putative
Length=821
Score = 65.5 bits (158), Expect = 7e-11, Method: Composition-based stats.
Identities = 38/119 (31%), Positives = 63/119 (52%), Gaps = 1/119 (0%)
Query 37 ETETLIVGGRVPKDVSDPDSMAEWSDTVEELPMPVKITIQSLSYLLPEEKKEAFEKASRF 96
E ET+I+GG D +DP++ +W++++ E PMP+K + LS +LP + +E+A RF
Sbjct 501 EKETVIIGGTTIFDPNDPNNFEKWAESISENPMPIKGEYEPLSRILPTRLSKIYEEALRF 560
Query 97 YAEA-VGMSRADLNAMGGKVESIGEVLRKATQVTYAGDPPGYAFCAPHERVLLGFALQL 154
Y V + + + +I E L KAT + +G C + LLGF+L +
Sbjct 561 YISVNVPSNFGQITDNEIRQYNIKEELMKATMIHSSGTGLVVVECEEKQNFLLGFSLSI 619
> tpv:TP04_0337 hypothetical protein
Length=498
Score = 51.2 bits (121), Expect = 1e-06, Method: Composition-based stats.
Identities = 32/112 (28%), Positives = 56/112 (50%), Gaps = 3/112 (2%)
Query 42 IVGGRVPKDVSDPDSMAEWSDTVEELPMPVKITIQSLSYLLPEEKKEAFEKASRFYAEAV 101
++GG++P D + A W++TV E PMP+ + SL L+ +++++A YA
Sbjct 272 VLGGKMPNFPMDDNEFAHWAETVAENPMPIGVVSTSLKTLMHPAMHQSYDQALHQYAILN 331
Query 102 GMSRADLNAMGGK-VESIGEVLRKATQVTYAGDPPGYAFCAPHERVLLGFAL 152
G+S L + G +E E+ T V++ D C +VL+G +L
Sbjct 332 GISYEKLKMISGNSIELSKEIENGTTIVSWNRD--NKIKCPRGSKVLMGISL 381
> tpv:TP03_0810 hypothetical protein
Length=348
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 29/106 (27%), Positives = 52/106 (49%), Gaps = 5/106 (4%)
Query 4 SALKATFGVVSGGVSSNVSKEAEQREQLKHFQYETETLIVGGRVPKDVS--DPDSMAEWS 61
S +K T +G SS S+ +R +K + + ++GG ++ DP ++W
Sbjct 245 SDIKNTKAKRTGEASSEFSEIFRRR--IKGNKIKKHLWVIGGSFVNNLEQIDPKMFSKWV 302
Query 62 DTVEELPMPVKITIQSLSYLLPEEKKEAFEKASRFYAEAVGMSRAD 107
T+++ PMP+K LS P EK E + KA +Y + G+ + +
Sbjct 303 KTIDKRPMPIKARFSELSMFFP-EKHETYMKAVEYYEQISGIKKLN 347
> bbo:BBOV_III000410 hypothetical protein
Length=1272
Score = 45.4 bits (106), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 27/96 (28%), Positives = 48/96 (50%), Gaps = 8/96 (8%)
Query 10 FGVVSG-GVSSNVSKEAEQREQLKHFQYETETLIVGGRVPKDVSDPDSMAEWSDTVEELP 68
FG +G G+S +K+ + + ++GGR +V D + EW +++ E P
Sbjct 1172 FGASAGVGLSKGSTKDNTENISFTYVN------VLGGRTIGNVEDENEYLEWINSIPEHP 1225
Query 69 MPVKITIQSLSYLL-PEEKKEAFEKASRFYAEAVGM 103
MP++ + L+ L EE KE ++ A +Y E G+
Sbjct 1226 MPIRNQLAPLAKLFDSEELKETYDDAMEYYVELNGL 1261
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/104 (25%), Positives = 50/104 (48%), Gaps = 2/104 (1%)
Query 54 PDSMAEWSDTVEELPMPVKITIQSLSYLLPEEK-KEAFEKASRFYAEAVGMSRADLNAMG 112
PD +W V P+P+ + Q LS ++ E + K + KA +Y + G+ + +
Sbjct 656 PDVFEDWVRDVALNPVPIDVEYQPLSEIMMENQAKNLYAKAIEYYTKLKGIDIKEKVSTE 715
Query 113 GKVESIGEVLRKATQVTYAGDPP-GYAFCAPHERVLLGFALQLN 155
I +L++ T V G+P C+ + ++++GF + LN
Sbjct 716 TIESPISHLLKELTMVAVDGNPRVNQDICSGNSKIVVGFGVNLN 759
Score = 34.7 bits (78), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 27/112 (24%), Positives = 54/112 (48%), Gaps = 6/112 (5%)
Query 3 KSALKATFGVVSGGVSSNVSKEAEQREQLKHFQYETETLIVGGRVPKDVSDPDSMAEWSD 62
K + GV+SG S N++ E++++ K + + I+GG V P+ +W D
Sbjct 331 KHNISVNVGVLSG--SFNLNNTNEKQQKEKVKKKNGKFFIMGGDTFIPVDTPEGFRDWVD 388
Query 63 TVEELPMPVKITIQSLSYLLPEEKKEAFEKASRFYA----EAVGMSRADLNA 110
+++E MP+ + + +S +P + + A + Y+ E+ G S + A
Sbjct 389 SIKENSMPINVELTPMSQFMPIGIRRHYWFALKSYSGGTDESTGFSLKNYRA 440
> bbo:BBOV_III000320 17.m10445; hypothetical protein
Length=512
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 31/116 (26%), Positives = 54/116 (46%), Gaps = 10/116 (8%)
Query 39 ETLIVGGRVPKDVSDPDSMA--EWSDTVEELPMPVKITIQSLSYLLPEEKKEAFEKASRF 96
+ LI+GG + DS A WS +V E PMP++ SL + + +K ++++ A RF
Sbjct 235 QMLILGGYYFSGLESNDSKAFNRWSKSVWERPMPIRANFTSLEHFMG-DKSKSYQHAVRF 293
Query 97 YAEAVGMSRADLNAMGGKVESIGEVLRKATQVTYAGDPPGYAFCAPHERVLLGFAL 152
Y ++ N +I ++L++ V G A+C V+ GF +
Sbjct 294 YRGVQNVA----NGHSLNYTTISQMLKETVTVV---SNTGSAYCPTGMSVVSGFVM 342
> bbo:BBOV_II001970 18.m09950; mac/perforin domain containing
membrane protein
Length=559
Score = 43.9 bits (102), Expect = 2e-04, Method: Composition-based stats.
Identities = 38/157 (24%), Positives = 65/157 (41%), Gaps = 24/157 (15%)
Query 1 NVKSALKATFGVVSGGV-SSNVSKEAEQREQLKHFQYETETLIVGGRVPKDVSDPDSMAE 59
NV A+KA V+SG + N +EQ++ +K Q DS+ +
Sbjct 278 NVDVAVKA---VISGALLEVNEKLNSEQQKAIKELQ------------------DDSLLK 316
Query 60 WSDTVEELPMPVKITIQSLSYLLPEEKKEAFEKASRFYAEAVGMSRADLNAMGGKVESIG 119
W TV PMP++ L + KEA+ A FY + G ++ G ++
Sbjct 317 WKSTVPIFPMPIRTIYAPLDMFIHSSYKEAYRNALNFYIKLNGALPWVVHQHNGTTLNVE 376
Query 120 EVLRKATQVTYAG-DPPGYAFCAPH-ERVLLGFALQL 154
++ +T + D PH ++V+ GF L++
Sbjct 377 SIINMSTALVLVNTDDDIVKLDCPHNDKVIFGFILEM 413
> tpv:TP03_0799 hypothetical protein
Length=356
Score = 42.0 bits (97), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 23/94 (24%), Positives = 46/94 (48%), Gaps = 2/94 (2%)
Query 37 ETETLIVGGRVPKDVSDPDSMAEWSDTVEELPMPVKITIQSLSYLLPEEKKEAFEKASRF 96
E TL +G P +VS+ +++W + V P P+ + + + ++PE+ + +E A ++
Sbjct 265 ELYTLTIGPEPPGNVSNSKVISDWLEKVVHNPTPIDLELVPIKQIIPEKYLKIYENALKY 324
Query 97 YAEAVGMSRADLNAMGGKVESIGEVLRKATQVTY 130
Y E + DLN++ I V + + Y
Sbjct 325 YQELNNI--KDLNSVNAVNSVINTVSLQLFYIVY 356
> bbo:BBOV_II002020 18.m06160; mac/perforin domain containing
protein
Length=420
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 25/88 (28%), Positives = 42/88 (47%), Gaps = 2/88 (2%)
Query 42 IVGGRVPKDVSDPDSMAEWSDTVEELPMPVKITIQSLSYLLPEEKKEAFEKASRFYAEAV 101
++GG +P A W ++V E PMP+ I SL L+ + ++++ A YAE
Sbjct 316 VIGGNMPNTPITDAEYAIWGNSVAENPMPIGIVGDSLKNLMDKNLRDSYSLAIHKYAELN 375
Query 102 GMSRADLNAM--GGKVESIGEVLRKATQ 127
G + L + GG+ + E RK +
Sbjct 376 GATYETLMKLENGGRTGLMKENARKVKK 403
> tpv:TP02_0166 hypothetical protein
Length=812
Score = 35.8 bits (81), Expect = 0.052, Method: Composition-based stats.
Identities = 32/139 (23%), Positives = 61/139 (43%), Gaps = 6/139 (4%)
Query 17 VSSNVSKEAEQREQLKHFQYETE--TLIVGGRVPKDVSDPDSMAEWSDTVEELPMPVKIT 74
+ SNV + + F +T+ T ++GG + AEW+ +V + MP+K
Sbjct 585 IDSNVEVSTIKSKNAGDFLLDTKKSTFVLGGDIYGH-GKTIEFAEWARSVADHAMPIKAE 643
Query 75 IQSLSYLLPEEKKEAFEKASRFYAEAVGMSRADLNAMGGKVESIGEVLRKATQVTYA-GD 133
+S+ + + ++A+ KA +Y + + +D +VE E L ++ +A G
Sbjct 644 FTPISHFIDKNLRDAYNKAYLYYGKVI--LESDFLRYQSQVELTPEDLISEVEIEHAQGT 701
Query 134 PPGYAFCAPHERVLLGFAL 152
A C + V GF +
Sbjct 702 GDVKASCKDGKLVQFGFMI 720
> pfa:PF08_0050 MAC/Perforin, putative
Length=654
Score = 35.8 bits (81), Expect = 0.052, Method: Composition-based stats.
Identities = 18/70 (25%), Positives = 36/70 (51%), Gaps = 4/70 (5%)
Query 60 WSDTVEELPMPVKITIQSLSYLLPEEKKEAFEKASRFYAEAVGMSRADLNAMGGKVESIG 119
W +++ +P+ + + SL+ +P EKKE+++ A FY G+ + + I
Sbjct 333 WKNSIYSNIVPIHLDLYSLNTFMPIEKKESYDMALLFYNNLYGIDNENFYLS----QDIT 388
Query 120 EVLRKATQVT 129
+VL + Q+T
Sbjct 389 DVLSEGKQIT 398
> tpv:TP03_0801 hypothetical protein
Length=353
Score = 35.8 bits (81), Expect = 0.066, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 35/65 (53%), Gaps = 4/65 (6%)
Query 34 FQYETETLIVGGRVPKDVSDPDSMAEWSDTVEELPMPVKITIQSLSYLLPEEK-KEAFEK 92
F+Y T I+GG D+S +W +V + PMP++ +S + + K+++++
Sbjct 283 FKY---TNILGGLPVSDISKESEYVKWIKSVYKYPMPIRTQFAPISKIFKSKALKDSYDE 339
Query 93 ASRFY 97
A RFY
Sbjct 340 AFRFY 344
> dre:100007189 Perforin-1-like
Length=574
Score = 32.3 bits (72), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 29/115 (25%), Positives = 56/115 (48%), Gaps = 22/115 (19%)
Query 4 SALKATFGVVSGGVSSNVSKEAEQR--EQLKH-----------FQYETETLIVGGRVPKD 50
+A+K V + G+ ++ EAE R +LK F E +T I+GG + +
Sbjct 242 TAIKDCLDVEASGIYKALTVEAEARLCRELKQKLGTNLKMSSKFS-ERQTEIIGGNINGE 300
Query 51 ------VSDPDSMAEWSDTVEELPMPVKITIQSLSYLLPEEK--KEAFEKASRFY 97
S P+++ W ++++ +P V+ T++ L +LL E+ ++ +KA Y
Sbjct 301 DLLFSGSSHPNALKGWLESLKSVPDVVQYTLKPLHFLLSEKHPARKGLKKAVEEY 355
> bbo:BBOV_II007150 18.m06592; mac/perforin domain containing
protein
Length=752
Score = 32.3 bits (72), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 23/99 (23%), Positives = 42/99 (42%), Gaps = 1/99 (1%)
Query 1 NVKSALKATFGVVSGGVSSNVSKEAEQREQLKHFQYETETLIVGGRVPKDVSDPDSMAEW 60
+VK+ L V + S N S + + + ++GG + D + +W
Sbjct 649 DVKAELSVQAEVATVDASVNASTSSLHNRDTESLDTKKSMFVIGGDIYGD-GNTIVFNDW 707
Query 61 SDTVEELPMPVKITIQSLSYLLPEEKKEAFEKASRFYAE 99
+ TV E MP+K L+ ++ E +A+ A FY +
Sbjct 708 AATVPENSMPIKAEYTPLAMIMGHEYMKAYNDAYLFYGK 746
> hsa:80725 SRCIN1, KIAA1684, P140, SNIP; SRC kinase signaling
inhibitor 1
Length=1183
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 18/36 (50%), Gaps = 0/36 (0%)
Query 115 VESIGEVLRKATQVTYAGDPPGYAFCAPHERVLLGF 150
ES G R A + AG PP F P ER L+GF
Sbjct 520 AESPGGKTRSAGSASTAGAPPSELFPGPGERSLVGF 555
> xla:379504 c9, MGC64276; complement component 9; K04000 complement
component 9
Length=593
Score = 30.0 bits (66), Expect = 3.3, Method: Composition-based stats.
Identities = 18/93 (19%), Positives = 40/93 (43%), Gaps = 17/93 (18%)
Query 1 NVKSALKATFGVVSGGVSSNVSKEAEQREQLKHFQYETETLIVGGRVPKDVSDPDSMAEW 60
N+K +++ V GG V+ E+ + + V+D + +W
Sbjct 407 NIKPVIESIISFVDGGTVEYVTALEEKLNK-----------------KQPVADVNDYVQW 449
Query 61 SDTVEELPMPVKITIQSLSYLLPEEKKEAFEKA 93
+ +++E P +K + L+P + K+A+ K+
Sbjct 450 ASSLKEAPAVIKSKPNPIYSLIPTDIKDAYTKS 482
> xla:100158389 hypothetical protein LOC100158389
Length=352
Score = 29.6 bits (65), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 11/29 (37%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 63 TVEELPMPVKITIQSLSYLLPEEKKEAFE 91
++E+L +P+ I+ QSL +++PE FE
Sbjct 46 SLEDLKIPIGISAQSLGHIIPETCNAIFE 74
> mmu:56013 Srcin1, P140, SNIP, mKIAA1684, p140Cap; SRC kinase
signaling inhibitor 1
Length=1217
Score = 29.3 bits (64), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 17/36 (47%), Gaps = 0/36 (0%)
Query 115 VESIGEVLRKATQVTYAGDPPGYAFCAPHERVLLGF 150
ES G R + AG PP F P ER L+GF
Sbjct 553 AESPGGKARSTGSASTAGAPPSELFPGPGERSLVGF 588
> eco:b0695 kdpD, ECK0683, JW0683, kac; fused sensory histidine
kinase in two-component regulatory system with KdpE: signal
sensing protein; K07646 two-component system, OmpR family,
sensor histidine kinase KdpD [EC:2.7.13.3]
Length=894
Score = 29.3 bits (64), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 27/51 (52%), Gaps = 3/51 (5%)
Query 5 ALKATFGVVSGGVSSNV---SKEAEQREQLKHFQYETETLIVGGRVPKDVS 52
A+ T G+V G +++ V ++ A REQ YE + GR P+D++
Sbjct 482 AVMLTVGLVIGNLTAGVRYQARVARYREQRTRHLYEMSKALAVGRSPQDIA 532
> dre:100007241 Perforin-1-like
Length=1012
Score = 28.5 bits (62), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 27/115 (23%), Positives = 56/115 (48%), Gaps = 22/115 (19%)
Query 4 SALKATFGVVSGGVSSNVSKEAEQR--EQLKH-----------FQYETETLIVGGRVPKD 50
+A+K V + G+ ++ EAE R +LK F E +T I+GG++ +
Sbjct 680 TAIKDCLDVEASGIYKALTVEAEARLCRELKQKLGTNLKMSSKFS-ERQTEIIGGKINGE 738
Query 51 ------VSDPDSMAEWSDTVEELPMPVKITIQSLSYLLPEEK--KEAFEKASRFY 97
S +++ W ++++ +P V+ T++ L +LL ++ ++ +KA Y
Sbjct 739 DLLFSGSSHKNALKGWLESLKSVPDVVQYTLKPLHFLLSDKHPARKGLKKAVEEY 793
Lambda K H
0.313 0.130 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3451799900
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40