bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1602_orf1
Length=136
Score E
Sequences producing significant alignments: (Bits) Value
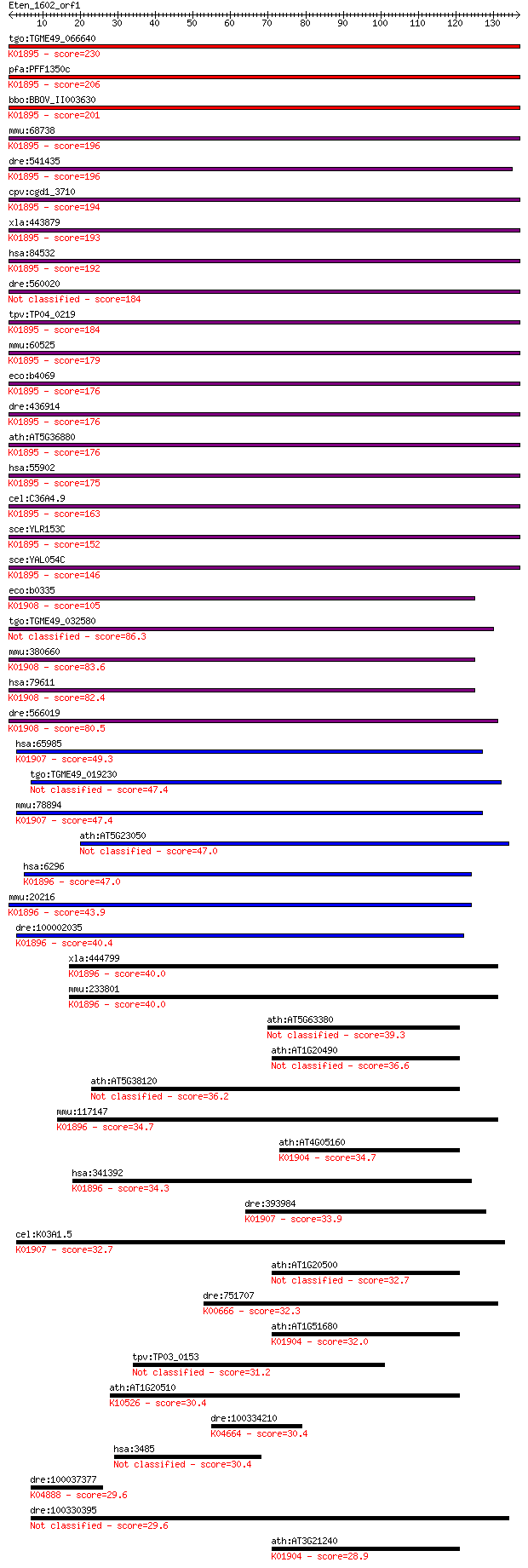
tgo:TGME49_066640 acetyl-coenzyme A synthetase, putative (EC:6... 230 1e-60
pfa:PFF1350c acetyl-CoA synthetase (EC:6.2.1.1); K01895 acetyl... 206 2e-53
bbo:BBOV_II003630 18.m06304; acetyl-CoA synthetase (EC:6.2.1.1... 201 4e-52
mmu:68738 Acss1, 1110032O15Rik, AI788978, Acas2, Acas2l, AceCS... 196 2e-50
dre:541435 acss1, MGC158738, MGC174587, cb395, fi28d10, sb:cb3... 196 3e-50
cpv:cgd1_3710 acetyl-coenzyme A synthetase ; K01895 acetyl-CoA... 194 1e-49
xla:443879 acss2.2, MGC80104, acas2, acecs, acs, acsa, acss2; ... 193 1e-49
hsa:84532 ACSS1, ACAS2L, AceCS2L, FLJ45659, MGC33843; acyl-CoA... 192 3e-49
dre:560020 hypothetical LOC560020 184 7e-47
tpv:TP04_0219 acetyl-coenzyme A synthetase (EC:6.2.1.1); K0189... 184 9e-47
mmu:60525 Acss2, 1110017C11Rik, ACAS, Acas1, Acas2, AceCS1, Ac... 179 2e-45
eco:b4069 acs, acsA, ECK4062, JW4030, yfaC; acetyl-CoA synthet... 176 1e-44
dre:436914 acss2, wu:fa04c03, wu:fj80b06, wu:fj80h04, zgc:9220... 176 2e-44
ath:AT5G36880 acetyl-CoA synthetase, putative / acetate-CoA li... 176 2e-44
hsa:55902 ACSS2, ACAS2, ACECS, ACS, ACSA, DKFZp762G026, dJ1161... 175 3e-44
cel:C36A4.9 hypothetical protein; K01895 acetyl-CoA synthetase... 163 1e-40
sce:YLR153C ACS2; Acetyl-coA synthetase isoform which, along w... 152 2e-37
sce:YAL054C ACS1, FUN44; Acetyl-coA synthetase isoform which, ... 146 2e-35
eco:b0335 prpE, ECK0332, JW0326, yahU; propionate--CoA ligase;... 105 5e-23
tgo:TGME49_032580 acetyl-coenzyme A synthetase, putative (EC:6... 86.3 2e-17
mmu:380660 Acss3, 8430416H19Rik, Gm874; acyl-CoA synthetase sh... 83.6 2e-16
hsa:79611 ACSS3, FLJ21963; acyl-CoA synthetase short-chain fam... 82.4 4e-16
dre:566019 acss3, si:dkey-231a18.1; acyl-CoA synthetase short-... 80.5 1e-15
hsa:65985 AACS, ACSF1, FLJ12389, FLJ41251, SUR-5; acetoacetyl-... 49.3 3e-06
tgo:TGME49_019230 acetoacetyl-CoA synthetase, putative (EC:6.2... 47.4 1e-05
mmu:78894 Aacs, 2210408B16Rik, SUR5; acetoacetyl-CoA synthetas... 47.4 1e-05
ath:AT5G23050 AAE17; AAE17 (ACYL-ACTIVATING ENZYME 17); cataly... 47.0 2e-05
hsa:6296 ACSM3, SA, SAH; acyl-CoA synthetase medium-chain fami... 47.0 2e-05
mmu:20216 Acsm3, Sa, Sah; acyl-CoA synthetase medium-chain fam... 43.9 2e-04
dre:100002035 acsm3, zgc:172040; acyl-CoA synthetase medium-ch... 40.4 0.002
xla:444799 acsm3, MGC82117; acyl-CoA synthetase medium-chain f... 40.0 0.002
mmu:233801 Acsm4, MGC56918, O-MACS, OMACS; acyl-CoA synthetase... 40.0 0.002
ath:AT5G63380 4-coumarate--CoA ligase family protein / 4-couma... 39.3 0.003
ath:AT1G20490 AMP-dependent synthetase and ligase family protein 36.6 0.026
ath:AT5G38120 4-coumarate--CoA ligase family protein / 4-couma... 36.2 0.028
mmu:117147 Acsm1, Acas3, Bucs1, Macs; acyl-CoA synthetase medi... 34.7 0.098
ath:AT4G05160 4-coumarate--CoA ligase, putative / 4-coumaroyl-... 34.7 0.100
hsa:341392 ACSM4; acyl-CoA synthetase medium-chain family memb... 34.3 0.10
dre:393984 aacs, MGC56105, zgc:56105; acetoacetyl-CoA syntheta... 33.9 0.14
cel:K03A1.5 sur-5; SUppressor of activated let-60 Ras family m... 32.7 0.31
ath:AT1G20500 4-coumarate-CoA ligase 32.7 0.32
dre:751707 acsf2, MGC152887, im:7145554, wu:fi49b09, zgc:15288... 32.3 0.48
ath:AT1G51680 4CL1; 4CL1 (4-COUMARATE:COA LIGASE 1); 4-coumara... 32.0 0.51
tpv:TP03_0153 hypothetical protein 31.2 1.0
ath:AT1G20510 OPCL1; OPCL1 (OPC-8:0 COA LIGASE1); 4-coumarate-... 30.4 1.5
dre:100334210 contact-like; K04664 growth differentiation fact... 30.4 1.5
hsa:3485 IGFBP2, IBP2, IGF-BP53; insulin-like growth factor bi... 30.4 1.9
dre:100037377 kcnc2, zgc:162956; potassium voltage-gated chann... 29.6 2.5
dre:100330395 EF-hand calcium binding domain 4B-like 29.6 3.2
ath:AT3G21240 4CL2; 4CL2 (4-COUMARATE:COA LIGASE 2); 4-coumara... 28.9 4.4
> tgo:TGME49_066640 acetyl-coenzyme A synthetase, putative (EC:6.2.1.1);
K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=535
Score = 230 bits (586), Expect = 1e-60, Method: Compositional matrix adjust.
Identities = 97/136 (71%), Positives = 122/136 (89%), Gaps = 0/136 (0%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
+CH+TAGYLLY+A TH+ +FDYHPGD + C+ADCGWITGHSY VYGPLCNGATT+LFQ I
Sbjct 151 LCHSTAGYLLYAALTHKCVFDYHPGDIYACVADCGWITGHSYVVYGPLCNGATTLLFQSI 210
Query 61 PTYPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
PTYP PGRYW++++KWK +Q YT+PTA+RALM++G+ WPKKY+LS+LRV+G+VGEPINPE
Sbjct 211 PTYPDPGRYWQMIEKWKASQFYTAPTALRALMRYGDSWPKKYELSSLRVLGTVGEPINPE 270
Query 121 AWRWYFTNVGRGHCSV 136
AWRWY + +G+ CSV
Sbjct 271 AWRWYASVIGQNRCSV 286
> pfa:PFF1350c acetyl-CoA synthetase (EC:6.2.1.1); K01895 acetyl-CoA
synthetase [EC:6.2.1.1]
Length=997
Score = 206 bits (523), Expect = 2e-53, Method: Composition-based stats.
Identities = 84/136 (61%), Positives = 107/136 (78%), Gaps = 0/136 (0%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
V HTTAGYLLY+ T +YIFD D FGC+AD GW+TGH+Y +YGPL NG TT++F I
Sbjct 566 VAHTTAGYLLYAYTTCKYIFDVKENDIFGCVADIGWVTGHTYVLYGPLLNGITTVIFSSI 625
Query 61 PTYPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
PTYP GRYW ++ KVTQ YT+PTA+RALM++G+EW +KYDLS+ R++GSVGEPINPE
Sbjct 626 PTYPDCGRYWSLIQTHKVTQFYTAPTALRALMKYGDEWIQKYDLSSCRILGSVGEPINPE 685
Query 121 AWRWYFTNVGRGHCSV 136
WRWY+ VG+ C++
Sbjct 686 TWRWYYNVVGKKKCTI 701
> bbo:BBOV_II003630 18.m06304; acetyl-CoA synthetase (EC:6.2.1.1);
K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=703
Score = 201 bits (512), Expect = 4e-52, Method: Compositional matrix adjust.
Identities = 85/136 (62%), Positives = 112/136 (82%), Gaps = 0/136 (0%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
V HTT GYL+Y+ T +YIFD H GD FGC+AD GWITGH+Y VYGPL NG TT +F +
Sbjct 317 VSHTTGGYLVYAHATTKYIFDAHVGDIFGCVADLGWITGHTYVVYGPLLNGLTTFMFSSL 376
Query 61 PTYPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
P YP PGRYW ++++ ++TQ YT+PTAIR+LM+HG+++P++YD+S++RV+GSVGEPINPE
Sbjct 377 PNYPDPGRYWRMIEQHRITQFYTAPTAIRSLMRHGDDYPRQYDISSVRVLGSVGEPINPE 436
Query 121 AWRWYFTNVGRGHCSV 136
AWRWY+ VGRG +V
Sbjct 437 AWRWYYEVVGRGRTNV 452
> mmu:68738 Acss1, 1110032O15Rik, AI788978, Acas2, Acas2l, AceCS2;
acyl-CoA synthetase short-chain family member 1 (EC:6.2.1.1);
K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=682
Score = 196 bits (497), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 85/136 (62%), Positives = 104/136 (76%), Gaps = 0/136 (0%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
+ HT AGYLLY+A TH+ +FDY PGD FGC+AD GWITGHSY VYGPLCNGATT+LF+
Sbjct 297 LVHTQAGYLLYAAMTHKLVFDYQPGDVFGCVADIGWITGHSYVVYGPLCNGATTVLFEST 356
Query 61 PTYPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
P YP GRYWE V + K+ Q Y +PTA+R L+++G+ W KKYD S+LR +GSVGEPIN E
Sbjct 357 PVYPDAGRYWETVQRLKINQFYGAPTAVRLLLKYGDAWVKKYDRSSLRTLGSVGEPINHE 416
Query 121 AWRWYFTNVGRGHCSV 136
AW W VG G C++
Sbjct 417 AWEWLHKVVGDGRCTL 432
> dre:541435 acss1, MGC158738, MGC174587, cb395, fi28d10, sb:cb395,
wu:fa05a10, wu:fc49f03, wu:fi28d10, zgc:113194, zgc:158738;
acyl-CoA synthetase short-chain family member 1 (EC:6.2.1.13);
K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=693
Score = 196 bits (497), Expect = 3e-50, Method: Compositional matrix adjust.
Identities = 83/134 (61%), Positives = 104/134 (77%), Gaps = 0/134 (0%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
+ HT AGYLLY++ TH+Y+FDY PGD FGC+AD GWITGHSY VYGPLCNGAT++LF+
Sbjct 308 IVHTQAGYLLYASLTHQYVFDYTPGDVFGCVADIGWITGHSYVVYGPLCNGATSVLFEST 367
Query 61 PTYPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
P YP+PGRYWE V + ++ Q Y +PTAIR L+++ E W KKYD S+L+ + SVGEPIN E
Sbjct 368 PVYPNPGRYWETVQRLRINQFYGAPTAIRLLLKYEESWVKKYDRSSLKTLASVGEPINHE 427
Query 121 AWRWYFTNVGRGHC 134
AW W+ VG G C
Sbjct 428 AWDWFHNVVGDGRC 441
> cpv:cgd1_3710 acetyl-coenzyme A synthetase ; K01895 acetyl-CoA
synthetase [EC:6.2.1.1]
Length=695
Score = 194 bits (492), Expect = 1e-49, Method: Compositional matrix adjust.
Identities = 84/136 (61%), Positives = 109/136 (80%), Gaps = 0/136 (0%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
V H+TAGYLLY+A T +Y+F+ HPGD FGC D GWITGHSY VY PLCNG TT++F+G+
Sbjct 310 VQHSTAGYLLYAAVTQKYLFNIHPGDIFGCAGDIGWITGHSYLVYAPLCNGITTLIFEGV 369
Query 61 PTYPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
PTYP+ GRYWE+V++ ++T Y +PTAIR L + G+++ KK+D S+LRV+GSVGEPINP
Sbjct 370 PTYPNAGRYWEMVERHRITHFYAAPTAIRTLKRLGDDFVKKHDRSSLRVLGSVGEPINPS 429
Query 121 AWRWYFTNVGRGHCSV 136
AWRWY + VG CS+
Sbjct 430 AWRWYHSVVGEERCSI 445
> xla:443879 acss2.2, MGC80104, acas2, acecs, acs, acsa, acss2;
acyl-CoA synthetase short-chain family member 2, gene 2 (EC:6.2.1.1);
K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=680
Score = 193 bits (490), Expect = 1e-49, Method: Compositional matrix adjust.
Identities = 85/136 (62%), Positives = 108/136 (79%), Gaps = 0/136 (0%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
V HT AGYLLY+A T +Y+FDYH D + AD GWITGHSY VYGPL NGAT+++F+GI
Sbjct 305 VVHTVAGYLLYTALTFKYVFDYHENDIYWSTADIGWITGHSYIVYGPLANGATSVMFEGI 364
Query 61 PTYPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
PTYPH GR+WEIV K+KVTQ YT+PTAIR LM++G E +K+DLS+L+++G+VGEPIN E
Sbjct 365 PTYPHVGRFWEIVQKYKVTQFYTAPTAIRLLMKYGNEAVQKFDLSSLKILGTVGEPINSE 424
Query 121 AWRWYFTNVGRGHCSV 136
AW WY+ +G C +
Sbjct 425 AWLWYYNVIGNQRCPI 440
> hsa:84532 ACSS1, ACAS2L, AceCS2L, FLJ45659, MGC33843; acyl-CoA
synthetase short-chain family member 1 (EC:6.2.1.1); K01895
acetyl-CoA synthetase [EC:6.2.1.1]
Length=689
Score = 192 bits (487), Expect = 3e-49, Method: Compositional matrix adjust.
Identities = 82/136 (60%), Positives = 105/136 (77%), Gaps = 0/136 (0%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
+ HT AGYLLY+A TH+ +FD+ PGD FGC+AD GWITGHSY VYGPLCNGAT++LF+
Sbjct 304 IVHTQAGYLLYAALTHKLVFDHQPGDIFGCVADIGWITGHSYVVYGPLCNGATSVLFEST 363
Query 61 PTYPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
P YP+ GRYWE V++ K+ Q Y +PTA+R L+++G+ W KKYD S+LR +GSVGEPIN E
Sbjct 364 PVYPNAGRYWETVERLKINQFYGAPTAVRLLLKYGDAWVKKYDRSSLRTLGSVGEPINCE 423
Query 121 AWRWYFTNVGRGHCSV 136
AW W VG C++
Sbjct 424 AWEWLHRVVGDSRCTL 439
> dre:560020 hypothetical LOC560020
Length=479
Score = 184 bits (467), Expect = 7e-47, Method: Compositional matrix adjust.
Identities = 81/136 (59%), Positives = 108/136 (79%), Gaps = 0/136 (0%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
V HT GY+LY++ T +Y+FDYH D + C AD GWITGHSY YGPL NGAT++LF+G+
Sbjct 286 VVHTVTGYMLYTSLTFKYVFDYHHDDVYWCTADIGWITGHSYITYGPLANGATSVLFEGV 345
Query 61 PTYPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
P +PH GR WEI++K+ V++ YT+PTAIR LM++G+E ++YDLS+LRV+G+VGEPINPE
Sbjct 346 PVHPHVGRLWEIIEKYGVSKFYTAPTAIRLLMKYGKEPLQRYDLSSLRVLGTVGEPINPE 405
Query 121 AWRWYFTNVGRGHCSV 136
AW WY+ VG+ C V
Sbjct 406 AWLWYYDVVGQQRCPV 421
> tpv:TP04_0219 acetyl-coenzyme A synthetase (EC:6.2.1.1); K01895
acetyl-CoA synthetase [EC:6.2.1.1]
Length=681
Score = 184 bits (466), Expect = 9e-47, Method: Compositional matrix adjust.
Identities = 83/136 (61%), Positives = 104/136 (76%), Gaps = 0/136 (0%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
+ HTT GYL+Y+ T YIFD H GD FGC+AD GWITGH+Y VYGPL NG TT++F +
Sbjct 302 LAHTTGGYLVYAYATVRYIFDSHEGDVFGCMADVGWITGHTYVVYGPLLNGITTVMFGSL 361
Query 61 PTYPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
P YP P RYW IV + +TQ YT+PTAIR+LM+ G+E K ++LS+LRV+GSVGEPINPE
Sbjct 362 PNYPTPDRYWNIVQEHGLTQFYTAPTAIRSLMKFGDEPLKGHNLSSLRVLGSVGEPINPE 421
Query 121 AWRWYFTNVGRGHCSV 136
AW+WY+ VG G +V
Sbjct 422 AWKWYYNVVGNGKVAV 437
> mmu:60525 Acss2, 1110017C11Rik, ACAS, Acas1, Acas2, AceCS1,
Acs1; acyl-CoA synthetase short-chain family member 2 (EC:6.2.1.1);
K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=701
Score = 179 bits (454), Expect = 2e-45, Method: Compositional matrix adjust.
Identities = 82/136 (60%), Positives = 102/136 (75%), Gaps = 0/136 (0%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
V HT GY+LY A T +Y+FD+HP D F C AD GWITGHSY YGPL NGAT++LF+GI
Sbjct 326 VVHTIGGYMLYVATTFKYVFDFHPEDVFWCTADIGWITGHSYVTYGPLANGATSVLFEGI 385
Query 61 PTYPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
PTYP GR W IVDK+KVT+ YT+PTAIR LM+ G++ K+ ++L+V+G+VGEPINPE
Sbjct 386 PTYPDEGRLWSIVDKYKVTKFYTAPTAIRMLMKFGDDPVTKHSRASLQVLGTVGEPINPE 445
Query 121 AWRWYFTNVGRGHCSV 136
AW WY VG C +
Sbjct 446 AWLWYHRVVGSQRCPI 461
> eco:b4069 acs, acsA, ECK4062, JW4030, yfaC; acetyl-CoA synthetase
(EC:6.2.1.1); K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=652
Score = 176 bits (447), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 79/136 (58%), Positives = 103/136 (75%), Gaps = 0/136 (0%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
V HTT GYL+Y+A T +Y+FDYHPGD + C AD GW+TGHSY +YGPL GATT++F+G+
Sbjct 274 VLHTTGGYLVYAALTFKYVFDYHPGDIYWCTADVGWVTGHSYLLYGPLACGATTLMFEGV 333
Query 61 PTYPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
P +P P R ++VDK +V LYT+PTAIRALM G++ + D S+LR++GSVGEPINPE
Sbjct 334 PNWPTPARMAQVVDKHQVNILYTAPTAIRALMAEGDKAIEGTDRSSLRILGSVGEPINPE 393
Query 121 AWRWYFTNVGRGHCSV 136
AW WY+ +G C V
Sbjct 394 AWEWYWKKIGNEKCPV 409
> dre:436914 acss2, wu:fa04c03, wu:fj80b06, wu:fj80h04, zgc:92200;
acyl-CoA synthetase short-chain family member 2 (EC:6.2.1.1);
K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=415
Score = 176 bits (446), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 78/136 (57%), Positives = 103/136 (75%), Gaps = 0/136 (0%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
V HT +GY+LY+A T + +FDYH D + C AD GWITGHSY YGPL NGAT++LF+G+
Sbjct 40 VLHTVSGYMLYTASTFKMVFDYHSDDVYWCTADIGWITGHSYITYGPLANGATSVLFEGL 99
Query 61 PTYPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
PTYP R WEIVDK+ V++ YT+PTAIR LM++G + KY ++L+++G+VGEPINPE
Sbjct 100 PTYPDVSRMWEIVDKYHVSKFYTAPTAIRLLMKYGSDPVHKYKRTSLKILGTVGEPINPE 159
Query 121 AWRWYFTNVGRGHCSV 136
AW+WY+ VG C V
Sbjct 160 AWQWYYNVVGEKRCPV 175
> ath:AT5G36880 acetyl-CoA synthetase, putative / acetate-CoA
ligase, putative (EC:6.2.1.1); K01895 acetyl-CoA synthetase
[EC:6.2.1.1]
Length=743
Score = 176 bits (445), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 75/136 (55%), Positives = 100/136 (73%), Gaps = 0/136 (0%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
V HTT GY++Y+A T +Y FDY D + C ADCGWITGHSY YGP+ NGAT ++F+G
Sbjct 370 VLHTTGGYMIYTATTFKYAFDYKSTDVYWCTADCGWITGHSYVTYGPMLNGATVVVFEGA 429
Query 61 PTYPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
P YP PGR W+IVDK+KV+ YT+PT +R+LM+ +++ ++ +LRV+GSVGEPINP
Sbjct 430 PNYPDPGRCWDIVDKYKVSIFYTAPTLVRSLMRDDDKFVTRHSRKSLRVLGSVGEPINPS 489
Query 121 AWRWYFTNVGRGHCSV 136
AWRW+F VG C +
Sbjct 490 AWRWFFNVVGDSRCPI 505
> hsa:55902 ACSS2, ACAS2, ACECS, ACS, ACSA, DKFZp762G026, dJ1161H23.1;
acyl-CoA synthetase short-chain family member 2 (EC:6.2.1.1);
K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=714
Score = 175 bits (444), Expect = 3e-44, Method: Compositional matrix adjust.
Identities = 81/136 (59%), Positives = 100/136 (73%), Gaps = 0/136 (0%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
V HT GY+LY A T +Y+FD+H D F C AD GWITGHSY YGPL NGAT++LF+GI
Sbjct 339 VVHTVGGYMLYVATTFKYVFDFHAEDVFWCTADIGWITGHSYVTYGPLANGATSVLFEGI 398
Query 61 PTYPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
PTYP R W IVDK+KVT+ YT+PTAIR LM+ G+E K+ ++L+V+G+VGEPINPE
Sbjct 399 PTYPDVNRLWSIVDKYKVTKFYTAPTAIRLLMKFGDEPVTKHSRASLQVLGTVGEPINPE 458
Query 121 AWRWYFTNVGRGHCSV 136
AW WY VG C +
Sbjct 459 AWLWYHRVVGAQRCPI 474
> cel:C36A4.9 hypothetical protein; K01895 acetyl-CoA synthetase
[EC:6.2.1.1]
Length=680
Score = 163 bits (413), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 74/136 (54%), Positives = 94/136 (69%), Gaps = 0/136 (0%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
+ HTTAGY+ Y+ T +Y FD D + C ADCGWITGHSY +YGPL NG I ++G+
Sbjct 305 IQHTTAGYMTYAYATTKYTFDAQEDDVYWCTADCGWITGHSYLLYGPLMNGLKGIWYEGV 364
Query 61 PTYPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
PTYP P R W++ DK+ VT+LYTSPTA RALM G +W + TL+VIG+VGEPINP
Sbjct 365 PTYPTPSRMWDVTDKYGVTKLYTSPTAARALMALGNQWLESSSRKTLKVIGTVGEPINPA 424
Query 121 AWRWYFTNVGRGHCSV 136
AW W + VG + S+
Sbjct 425 AWMWLYKQVGLSNVSI 440
> sce:YLR153C ACS2; Acetyl-coA synthetase isoform which, along
with Acs1p, is the nuclear source of acetyl-coA for histone
acetylation; mutants affect global transcription; required
for growth on glucose; expressed under anaerobic conditions
(EC:6.2.1.1); K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=683
Score = 152 bits (385), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 69/136 (50%), Positives = 88/136 (64%), Gaps = 0/136 (0%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
V HTT GYLL +A T Y+FD HP D D GWITGH+Y +YGPL G +I+F+
Sbjct 288 VVHTTGGYLLGAALTTRYVFDIHPEDVLFTAGDVGWITGHTYALYGPLTLGTASIIFEST 347
Query 61 PTYPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
P YP GRYW I+ + K T Y +PTA+R + + GE KYD S+LRV+GSVGEPI+P+
Sbjct 348 PAYPDYGRYWRIIQRHKATHFYVAPTALRLIKRVGEAEIAKYDTSSLRVLGSVGEPISPD 407
Query 121 AWRWYFTNVGRGHCSV 136
W WY VG +C +
Sbjct 408 LWEWYHEKVGNKNCVI 423
> sce:YAL054C ACS1, FUN44; Acetyl-coA synthetase isoform which,
along with Acs2p, is the nuclear source of acetyl-coA for
histone acetlyation; expressed during growth on nonfermentable
carbon sources and under aerobic conditions (EC:6.2.1.1);
K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=713
Score = 146 bits (368), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 66/136 (48%), Positives = 88/136 (64%), Gaps = 0/136 (0%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
V H+TAGYLL + T Y FD H D F D GWITGH+Y VYGPL G T++F+G
Sbjct 330 VQHSTAGYLLGALLTMRYTFDTHQEDVFFTAGDIGWITGHTYVVYGPLLYGCATLVFEGT 389
Query 61 PTYPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
P YP+ RYW+I+D+ KVTQ Y +PTA+R L + G+ + + + L +LR +GSVGEPI E
Sbjct 390 PAYPNYSRYWDIIDEHKVTQFYVAPTALRLLKRAGDSYIENHSLKSLRCLGSVGEPIAAE 449
Query 121 AWRWYFTNVGRGHCSV 136
W WY +G+ +
Sbjct 450 VWEWYSEKIGKNEIPI 465
> eco:b0335 prpE, ECK0332, JW0326, yahU; propionate--CoA ligase;
K01908 propionyl-CoA synthetase [EC:6.2.1.17]
Length=628
Score = 105 bits (261), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 49/124 (39%), Positives = 76/124 (61%), Gaps = 0/124 (0%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
V GY + A + + IF G F C +D GW+ GHSY VY PL G TI+++G+
Sbjct 250 VQRDVGGYAVALATSMDTIFGGKAGSVFFCASDIGWVVGHSYIVYAPLLAGMATIVYEGL 309
Query 61 PTYPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
PT+P G +W IV+K++V++++++PTAIR L + +K+DLS+L V+ GEP++
Sbjct 310 PTWPDCGVWWTIVEKYQVSRMFSAPTAIRVLKKFPTAEIRKHDLSSLEVLYLAGEPLDEP 369
Query 121 AWRW 124
W
Sbjct 370 TASW 373
> tgo:TGME49_032580 acetyl-coenzyme A synthetase, putative (EC:6.2.1.1)
Length=821
Score = 86.3 bits (212), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 48/133 (36%), Positives = 74/133 (55%), Gaps = 6/133 (4%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
+ AG + S T I+ PG+T C +D GW+ GHS VYG L G T+++++G
Sbjct 391 IVRDHAGQCVSSHYTASAIYGIRPGNTVFCASDLGWVLGHSIMVYGTLIGGGTSVMYEGK 450
Query 61 PTYPH-PGRYWEIVDKWKVTQLYTSPTAIRALM---QHGEEWPKKYDLSTLRVIGSVGEP 116
P G +W I ++ V++++T+P+A+R + HGE + YDLS LR + VGE
Sbjct 451 PVMAKDAGAFWRISSEYGVSKIFTAPSALRNIRVADPHGERL-RAYDLSRLRTVYLVGER 509
Query 117 INPEAWRWYFTNV 129
+ +RW FTN
Sbjct 510 TDHATFRW-FTNA 521
> mmu:380660 Acss3, 8430416H19Rik, Gm874; acyl-CoA synthetase
short-chain family member 3 (EC:6.2.1.1); K01908 propionyl-CoA
synthetase [EC:6.2.1.17]
Length=682
Score = 83.6 bits (205), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 46/127 (36%), Positives = 68/127 (53%), Gaps = 3/127 (2%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
V T GY + T I+ PG+ + +D GW+ GHSY YGPL +G TT+L++G
Sbjct 304 VVRPTGGYAVMLNWTMSSIYGLKPGEVWWAASDLGWVVGHSYICYGPLLHGNTTVLYEGK 363
Query 61 PT-YPHPGRYWEIVDKWKVTQLYTSPTAIRALMQH--GEEWPKKYDLSTLRVIGSVGEPI 117
P P G Y+ ++ + V L+T+PTAIRA+ Q G K+Y L+ + + GE
Sbjct 364 PVGTPDAGAYFRVLAEHGVAALFTAPTAIRAIRQQDPGAALGKQYSLTRFKTLFVAGERC 423
Query 118 NPEAWRW 124
+ E W
Sbjct 424 DVETLEW 430
> hsa:79611 ACSS3, FLJ21963; acyl-CoA synthetase short-chain family
member 3 (EC:6.2.1.1); K01908 propionyl-CoA synthetase
[EC:6.2.1.17]
Length=686
Score = 82.4 bits (202), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 45/127 (35%), Positives = 68/127 (53%), Gaps = 3/127 (2%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
V T GY + + I+ PG+ + +D GW+ GHSY YGPL +G TT+L++G
Sbjct 309 VIRPTGGYAVMLHWSMSSIYGLQPGEVWWAASDLGWVVGHSYICYGPLLHGNTTVLYEGK 368
Query 61 PT-YPHPGRYWEIVDKWKVTQLYTSPTAIRALMQH--GEEWPKKYDLSTLRVIGSVGEPI 117
P P G Y+ ++ + V L+T+PTAIRA+ Q G K+Y L+ + + GE
Sbjct 369 PVGTPDAGAYFRVLAEHGVAALFTAPTAIRAIRQQDPGAALGKQYSLTRFKTLFVAGERC 428
Query 118 NPEAWRW 124
+ E W
Sbjct 429 DVETLEW 435
> dre:566019 acss3, si:dkey-231a18.1; acyl-CoA synthetase short-chain
family member 3; K01908 propionyl-CoA synthetase [EC:6.2.1.17]
Length=714
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 45/133 (33%), Positives = 70/133 (52%), Gaps = 3/133 (2%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
V + GY + T ++ PG+ + +D GW+ GHSY YGPL +G TT+L++G
Sbjct 331 VVRDSGGYAVMLNWTMSNVYGLAPGEVWWAASDLGWVVGHSYICYGPLLHGNTTVLYEGK 390
Query 61 PT-YPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHG--EEWPKKYDLSTLRVIGSVGEPI 117
P P PG ++ ++ + ++T+PTAIRA+ Q E K Y L+ LR + GE
Sbjct 391 PVGTPDPGAFFRVMSEHGTACMFTAPTAIRAIRQQDPQAEHGKLYPLNRLRNLFVAGERC 450
Query 118 NPEAWRWYFTNVG 130
+ E W + G
Sbjct 451 DIETLEWAKKSFG 463
> hsa:65985 AACS, ACSF1, FLJ12389, FLJ41251, SUR-5; acetoacetyl-CoA
synthetase (EC:6.2.1.16); K01907 acetoacetyl-CoA synthetase
[EC:6.2.1.16]
Length=672
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 29/124 (23%), Positives = 52/124 (41%), Gaps = 2/124 (1%)
Query 3 HTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGIPT 62
H+ G L+ H + D C GW+ + + L GA +L+ G P
Sbjct 302 HSAGGTLIQHLKEHLLHGNMTSSDILLCYTTVGWMMWN--WMVSLLATGAAMVLYDGSPL 359
Query 63 YPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPEAW 122
P P W++VD+ +T L T + L + + + + L L I S G P+ +++
Sbjct 360 VPTPNVLWDLVDRIGITVLVTGAKWLSVLEEKAMKPVETHSLQMLHTILSTGSPLKAQSY 419
Query 123 RWYF 126
+ +
Sbjct 420 EYVY 423
> tgo:TGME49_019230 acetoacetyl-CoA synthetase, putative (EC:6.2.1.16)
Length=535
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 31/125 (24%), Positives = 58/125 (46%), Gaps = 2/125 (1%)
Query 7 GYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGIPTYPHP 66
G LL H+ + PGD+ + W+ + + V G L +GA+ +L++G YP P
Sbjct 140 GLLLQLVKEHQLHLNVRPGDSMLYYSTASWMMWN-WLVAG-LASGASLLLYEGHAMYPDP 197
Query 67 GRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPEAWRWYF 126
W D+ T TS ++ L + + ++ LR + S G P+ P +R+
Sbjct 198 LVLWRFFDQHGGTLFGTSAKYLQDLEKMDIVPKATFAMNRLRTLCSTGSPLYPHTFRYAA 257
Query 127 TNVGR 131
+++ +
Sbjct 258 SSIKK 262
> mmu:78894 Aacs, 2210408B16Rik, SUR5; acetoacetyl-CoA synthetase
(EC:6.2.1.16); K01907 acetoacetyl-CoA synthetase [EC:6.2.1.16]
Length=672
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 29/124 (23%), Positives = 54/124 (43%), Gaps = 2/124 (1%)
Query 3 HTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGIPT 62
H+ G L+ H + D GW+ + + L GA+ +L+ G P
Sbjct 302 HSAGGTLIQHLKEHMLHGNMTSSDILLYYTTVGWMMWN--WMVSALATGASLVLYDGSPL 359
Query 63 YPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPEAW 122
P P W++VD+ +T L T + L + + + ++L TL I S G P+ +++
Sbjct 360 VPTPNVLWDLVDRIGITILGTGAKWLSVLEEKDMKPVETHNLHTLHTILSTGSPLKAQSY 419
Query 123 RWYF 126
+ +
Sbjct 420 EYVY 423
> ath:AT5G23050 AAE17; AAE17 (ACYL-ACTIVATING ENZYME 17); catalytic/
ligase (EC:6.2.1.1)
Length=721
Score = 47.0 bits (110), Expect = 2e-05, Method: Composition-based stats.
Identities = 34/114 (29%), Positives = 48/114 (42%), Gaps = 9/114 (7%)
Query 20 FDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGIPTYPHPGRYWEIVDKWKVT 79
D GD + GW+ G + VY L NGA L+ G P P ++ V +V+
Sbjct 385 LDVQRGDVVAWPTNLGWMMG-PWLVYASLINGACMGLYNGSPLGPTFAKF---VQDAEVS 440
Query 80 QLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPEAWRWYFTNVGRGH 133
L P+ +R YD S +R GS GE N + + W +GR H
Sbjct 441 VLGVIPSIVRTWQNSNST--SGYDWSRIRCFGSTGEASNIDEYLWL---MGRAH 489
> hsa:6296 ACSM3, SA, SAH; acyl-CoA synthetase medium-chain family
member 3 (EC:6.2.1.2); K01896 medium-chain acyl-CoA synthetase
[EC:6.2.1.2]
Length=586
Score = 47.0 bits (110), Expect = 2e-05, Method: Composition-based stats.
Identities = 36/133 (27%), Positives = 55/133 (41%), Gaps = 19/133 (14%)
Query 5 TAGYLLYSAPTH-----------EYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGAT 53
T+GY +A TH + D P D +D GW +V+ P GA
Sbjct 238 TSGYPKMTAHTHSSFGLGLSVNGRFWLDLTPSDVMWNTSDTGWAKSAWSSVFSPWIQGA- 296
Query 54 TILFQGIPTYPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSV 113
+ +P + P + + K+ +T ++PT R L+Q+ Y +L+ S
Sbjct 297 CVFTHHLPRF-EPTSILQTLSKYPITVFCSAPTVYRMLVQND---ITSYKFKSLKHCVSA 352
Query 114 GEPINP---EAWR 123
GEPI P E WR
Sbjct 353 GEPITPDVTEKWR 365
> mmu:20216 Acsm3, Sa, Sah; acyl-CoA synthetase medium-chain family
member 3 (EC:6.2.1.2); K01896 medium-chain acyl-CoA synthetase
[EC:6.2.1.2]
Length=580
Score = 43.9 bits (102), Expect = 2e-04, Method: Composition-based stats.
Identities = 33/126 (26%), Positives = 55/126 (43%), Gaps = 8/126 (6%)
Query 1 VCHTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGI 60
+ HT + + L + + D D +D GW +V+ P GA + +
Sbjct 239 IGHTHSSFGLGLSVNGRFWLDLIASDVMWNTSDTGWAKSAWSSVFSPWTQGAC-VFAHYL 297
Query 61 PTYPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
P + + + K+ +T ++PTA R L+Q+ Y ++L+ S GEPINPE
Sbjct 298 PRF-ESTSILQTLSKFPITVFCSAPTAYRMLVQNDMS---SYKFNSLKHCVSAGEPINPE 353
Query 121 A---WR 123
WR
Sbjct 354 VMEQWR 359
> dre:100002035 acsm3, zgc:172040; acyl-CoA synthetase medium-chain
family member 3 (EC:6.2.1.2); K01896 medium-chain acyl-CoA
synthetase [EC:6.2.1.2]
Length=591
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 33/119 (27%), Positives = 46/119 (38%), Gaps = 5/119 (4%)
Query 3 HTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGIPT 62
H+ Y L Y D D F +D GW +V+ P GA + +P
Sbjct 250 HSHCSYGLGLTVNGRYWLDLTEQDVFWNTSDTGWAKSAWSSVFAPWIQGA-CVFVHHMPR 308
Query 63 YPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPEA 121
+ + + + +T T+PTA R L+Q KY L GEPINPE
Sbjct 309 F-DTNTVLKTLSHYPITTFCTAPTAYRMLVQDD---LSKYKFQALEHCLCAGEPINPEV 363
> xla:444799 acsm3, MGC82117; acyl-CoA synthetase medium-chain
family member 3; K01896 medium-chain acyl-CoA synthetase [EC:6.2.1.2]
Length=584
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/114 (23%), Positives = 47/114 (41%), Gaps = 5/114 (4%)
Query 17 EYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGIPTYPHPGRYWEIVDKW 76
+Y D P D ++D GW +V+ P G + + +P + P + + +
Sbjct 256 KYWMDLTPSDIVWNMSDTGWAKSAWSSVFAPWIQG-SCVFAHSMPRF-DPNIVLQTLSTF 313
Query 77 KVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPEAWRWYFTNVG 130
+T ++PTA R + Y +L+ S GEPINP+ + G
Sbjct 314 PITTFCSAPTAYRIFVLQNL---ASYQFKSLQHCVSAGEPINPQVMEQWKEQTG 364
> mmu:233801 Acsm4, MGC56918, O-MACS, OMACS; acyl-CoA synthetase
medium-chain family member 4 (EC:6.2.1.2); K01896 medium-chain
acyl-CoA synthetase [EC:6.2.1.2]
Length=580
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 33/125 (26%), Positives = 46/125 (36%), Gaps = 27/125 (21%)
Query 17 EYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGA-----------TTILFQGIPTYPH 65
Y D D ++D GWI +V+ GA T I + TYP
Sbjct 255 RYWLDLTSSDIMWNMSDTGWIKAAIGSVFSTWLRGACVFVHRMAQFDTDIFLDTLTTYP- 313
Query 66 PGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPEAWRWY 125
+T L ++PT R L+Q K+Y LR + GEP+NPE +
Sbjct 314 ------------ITTLCSAPTVYRMLVQKD---LKRYQFKRLRHCLTGGEPLNPEVLEQW 358
Query 126 FTNVG 130
G
Sbjct 359 KMQTG 363
> ath:AT5G63380 4-coumarate--CoA ligase family protein / 4-coumaroyl-CoA
synthase family protein
Length=562
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 21/51 (41%), Positives = 32/51 (62%), Gaps = 2/51 (3%)
Query 70 WEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
++ V+K+KVT + SP I AL++ E KKYDL +LR +G G P+ +
Sbjct 288 FKAVEKYKVTGMPVSPPLIVALVK--SELTKKYDLRSLRSLGCGGAPLGKD 336
> ath:AT1G20490 AMP-dependent synthetase and ligase family protein
Length=447
Score = 36.6 bits (83), Expect = 0.026, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 30/50 (60%), Gaps = 0/50 (0%)
Query 71 EIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
+ V+K+K T L +P + A++ ++ KYDL++LR + G P++ E
Sbjct 281 QAVEKYKATILSLAPPVLVAMINGADQLKAKYDLTSLRKVRCGGAPLSKE 330
> ath:AT5G38120 4-coumarate--CoA ligase family protein / 4-coumaroyl-CoA
synthase family protein
Length=550
Score = 36.2 bits (82), Expect = 0.028, Method: Composition-based stats.
Identities = 26/98 (26%), Positives = 42/98 (42%), Gaps = 4/98 (4%)
Query 23 HPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGIPTYPHPGRYWEIVDKWKVTQLY 82
P TF C G V L G T ++ +P + G V+K++ T L
Sbjct 238 QPQQTFICTVPLFHTFGLLNFVLATLALGTTVVI---LPRF-DLGEMMAAVEKYRATTLI 293
Query 83 TSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
P + ++ ++ KKYD+S LR + G P++ E
Sbjct 294 LVPPVLVTMINKADQIMKKYDVSFLRTVRCGGAPLSKE 331
> mmu:117147 Acsm1, Acas3, Bucs1, Macs; acyl-CoA synthetase medium-chain
family member 1 (EC:6.2.1.2); K01896 medium-chain
acyl-CoA synthetase [EC:6.2.1.2]
Length=573
Score = 34.7 bits (78), Expect = 0.098, Method: Compositional matrix adjust.
Identities = 26/117 (22%), Positives = 46/117 (39%), Gaps = 5/117 (4%)
Query 14 PTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGIPTYPHPGRYWEIV 73
P+ + D C++D GWI + P +G T+ +P + P E++
Sbjct 245 PSCRKLLKLKTSDILWCMSDPGWILATVGCLIEPWTSGC-TVFIHHLPQF-DPKVIVEVL 302
Query 74 DKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPEAWRWYFTNVG 130
K+ +TQ +P R ++Q TL + GE + PE + + G
Sbjct 303 FKYPITQCLAAPGVYRMVLQQKT---SNLRFPTLEHCTTGGESLLPEEYEQWKQRTG 356
> ath:AT4G05160 4-coumarate--CoA ligase, putative / 4-coumaroyl-CoA
synthase, putative; K01904 4-coumarate--CoA ligase [EC:6.2.1.12]
Length=544
Score = 34.7 bits (78), Expect = 0.100, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 29/48 (60%), Gaps = 2/48 (4%)
Query 73 VDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
++K++VT L+ P AL + + KK+DLS+L+ IGS P+ +
Sbjct 277 IEKFRVTHLWVVPPVFLALSK--QSIVKKFDLSSLKYIGSGAAPLGKD 322
> hsa:341392 ACSM4; acyl-CoA synthetase medium-chain family member
4 (EC:6.2.1.2); K01896 medium-chain acyl-CoA synthetase
[EC:6.2.1.2]
Length=580
Score = 34.3 bits (77), Expect = 0.10, Method: Composition-based stats.
Identities = 28/109 (25%), Positives = 45/109 (41%), Gaps = 8/109 (7%)
Query 18 YIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGIPTYPHPGRYWEIVDKWK 77
Y D D ++D GW+ +V+ GA + + + + +
Sbjct 256 YWLDLKSSDIIWNMSDTGWVKAAIGSVFSSWLCGACVFVHRMAQFDTDTFL--DTLTTYP 313
Query 78 VTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPEA---WR 123
+T L + PT R L+Q K+Y +LR + GEP+NPE WR
Sbjct 314 ITTLCSPPTVYRMLVQKDL---KRYKFKSLRHCLTGGEPLNPEVLEQWR 359
> dre:393984 aacs, MGC56105, zgc:56105; acetoacetyl-CoA synthetase
(EC:6.2.1.16); K01907 acetoacetyl-CoA synthetase [EC:6.2.1.16]
Length=659
Score = 33.9 bits (76), Expect = 0.14, Method: Composition-based stats.
Identities = 14/64 (21%), Positives = 31/64 (48%), Gaps = 0/64 (0%)
Query 64 PHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPEAWR 123
P W++VD+ +T T + L + ++ + L TL + S G P+ P+++
Sbjct 348 PSANVLWDLVDRLGITIFGTGAKWLAVLEERDQKPASTHSLQTLHTLLSTGSPLKPQSYE 407
Query 124 WYFT 127
+ ++
Sbjct 408 YVYS 411
> cel:K03A1.5 sur-5; SUppressor of activated let-60 Ras family
member (sur-5); K01907 acetoacetyl-CoA synthetase [EC:6.2.1.16]
Length=700
Score = 32.7 bits (73), Expect = 0.31, Method: Composition-based stats.
Identities = 29/130 (22%), Positives = 47/130 (36%), Gaps = 2/130 (1%)
Query 3 HTTAGYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGIPT 62
HT G LL H D D CGW+ + + L + + +LF P
Sbjct 331 HTVGGTLLKHIEEHLVQGDSKKHDRMFFYTTCGWMMYNWMISF--LYSKGSVVLFDECPL 388
Query 63 YPHPGRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPEAW 122
P +I K + T + ++ + YDLS + + S G P+ E +
Sbjct 389 APDTHIIMKIAAKTQSTMIGMGAKLYDEYLRLQIPFNTLYDLSKIHTVYSTGSPLKKECF 448
Query 123 RWYFTNVGRG 132
+ T + G
Sbjct 449 AYINTYIAPG 458
> ath:AT1G20500 4-coumarate-CoA ligase
Length=550
Score = 32.7 bits (73), Expect = 0.32, Method: Composition-based stats.
Identities = 15/50 (30%), Positives = 28/50 (56%), Gaps = 0/50 (0%)
Query 71 EIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
+ V+K + T L +P + A++ + KYDLS+L+ + G P++ E
Sbjct 279 DAVEKHRATALALAPPVLVAMINDADLIKAKYDLSSLKTVRCGGAPLSKE 328
> dre:751707 acsf2, MGC152887, im:7145554, wu:fi49b09, zgc:152887;
acyl-CoA synthetase family member 2 (EC:6.2.1.-); K00666
fatty-acyl-CoA synthase [EC:6.2.1.-]
Length=606
Score = 32.3 bits (72), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 40/79 (50%), Gaps = 6/79 (7%)
Query 53 TTILFQGIPTYPHPGRY-WEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIG 111
TT++F P+ + GR ++K K T +Y +PT ++ G+ K+DLS++R
Sbjct 318 TTVIF---PSTGYDGRANLRAIEKEKCTFVYGTPTMYIDML--GQPDLAKFDLSSVRGGI 372
Query 112 SVGEPINPEAWRWYFTNVG 130
+ G P PE R +G
Sbjct 373 AAGSPCPPEVMRKILNVMG 391
> ath:AT1G51680 4CL1; 4CL1 (4-COUMARATE:COA LIGASE 1); 4-coumarate-CoA
ligase (EC:6.2.1.12); K01904 4-coumarate--CoA ligase
[EC:6.2.1.12]
Length=539
Score = 32.0 bits (71), Expect = 0.51, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 28/50 (56%), Gaps = 2/50 (4%)
Query 71 EIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
E++ + KVT P + A+ + E +KYDLS++RV+ S P+ E
Sbjct 289 ELIQRCKVTVAPMVPPIVLAIAKSSET--EKYDLSSIRVVKSGAAPLGKE 336
> tpv:TP03_0153 hypothetical protein
Length=770
Score = 31.2 bits (69), Expect = 1.0, Method: Composition-based stats.
Identities = 24/77 (31%), Positives = 34/77 (44%), Gaps = 21/77 (27%)
Query 34 CGWITGHSYTVYGP----------LCNGATTILFQGIPTYPHPGRYWEIVDKWKVTQLYT 83
CG I+GHSYT GP L +GA L +G+ Y EI++K Y
Sbjct 475 CGEISGHSYTFLGPYGLSRKDIAYLKDGAKLTLTRGLVYYN------EIINKG-----YK 523
Query 84 SPTAIRALMQHGEEWPK 100
P + + G++ PK
Sbjct 524 DPEGDTSCPEDGDDKPK 540
> ath:AT1G20510 OPCL1; OPCL1 (OPC-8:0 COA LIGASE1); 4-coumarate-CoA
ligase; K10526 OPC-8:0 CoA ligase 1 [EC:6.2.1.-]
Length=546
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 22/93 (23%), Positives = 39/93 (41%), Gaps = 4/93 (4%)
Query 28 FGCLADCGWITGHSYTVYGPLCNGATTILFQGIPTYPHPGRYWEIVDKWKVTQLYTSPTA 87
F C I G + G L G+T I+ + + K++ T L P
Sbjct 234 FICTVPMFHIYGLAAFATGLLAYGSTIIVLSKFEMH----EMMSAIGKYQATSLPLVPPI 289
Query 88 IRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
+ A++ ++ KYDLS++ + G P++ E
Sbjct 290 LVAMVNGADQIKAKYDLSSMHTVLCGGAPLSKE 322
> dre:100334210 contact-like; K04664 growth differentiation factor
5/6/7
Length=474
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 11/24 (45%), Positives = 13/24 (54%), Gaps = 0/24 (0%)
Query 55 ILFQGIPTYPHPGRYWEIVDKWKV 78
+L Q P H YWE+ D WKV
Sbjct 251 VLLQARPFDSHSASYWEVFDIWKV 274
> hsa:3485 IGFBP2, IBP2, IGF-BP53; insulin-like growth factor
binding protein 2, 36kDa
Length=328
Score = 30.4 bits (67), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 19/39 (48%), Gaps = 7/39 (17%)
Query 29 GCLADCGWITGHSYTVYGPLCNGATTILFQGIPTYPHPG 67
GC + C + G + VY P C QG+ YPHPG
Sbjct 88 GCCSVCARLEGEACGVYTPRCG-------QGLRCYPHPG 119
> dre:100037377 kcnc2, zgc:162956; potassium voltage-gated channel,
Shaw-related subfamily, member 2; K04888 potassium voltage-gated
channel Shaw-related subfamily C member 2
Length=597
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 11/19 (57%), Positives = 15/19 (78%), Gaps = 0/19 (0%)
Query 7 GYLLYSAPTHEYIFDYHPG 25
G++ YSA ++EY FD HPG
Sbjct 55 GFIEYSARSNEYFFDRHPG 73
> dre:100330395 EF-hand calcium binding domain 4B-like
Length=403
Score = 29.6 bits (65), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 32/127 (25%), Positives = 50/127 (39%), Gaps = 14/127 (11%)
Query 7 GYLLYSAPTHEYIFDYHPGDTFGCLADCGWITGHSYTVYGPLCNGATTILFQGIPTYPHP 66
G L SA E +FD D G L + +G S ++GP +P PH
Sbjct 63 GELPLSADDLENVFDSLDADANGYLTFEEFSSGFSEFMFGPSV----------VPVDPHG 112
Query 67 GRYWEIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPEAWRWYF 126
G E+V + LY S R + GE+ +K+ + +G+ +PE R +
Sbjct 113 G--EELVSRKSPEMLYESQWEER--LSRGEDDEEKHFCMLMENLGASNIFEDPEEVRSLW 168
Query 127 TNVGRGH 133
+ R
Sbjct 169 VQLRRDE 175
> ath:AT3G21240 4CL2; 4CL2 (4-COUMARATE:COA LIGASE 2); 4-coumarate-CoA
ligase (EC:6.2.1.12); K01904 4-coumarate--CoA ligase
[EC:6.2.1.12]
Length=556
Score = 28.9 bits (63), Expect = 4.4, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 27/50 (54%), Gaps = 2/50 (4%)
Query 71 EIVDKWKVTQLYTSPTAIRALMQHGEEWPKKYDLSTLRVIGSVGEPINPE 120
E + + KVT P + A+ + E +KYDLS++R++ S P+ E
Sbjct 282 EQIQRCKVTVAMVVPPIVLAIAKSPET--EKYDLSSVRMVKSGAAPLGKE 329
Lambda K H
0.322 0.141 0.500
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2362384368
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40