bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1569_orf1
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
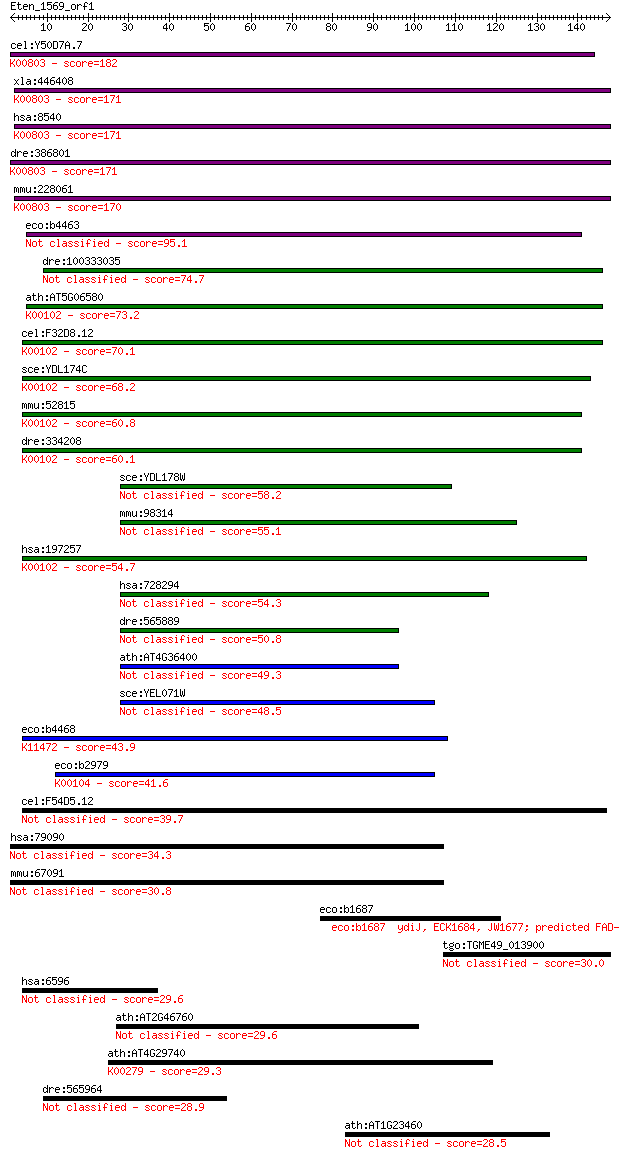
cel:Y50D7A.7 ads-1; Alkyl-Dihydroxyacetonephosphate Synthase f... 182 4e-46
xla:446408 agps, MGC83829; alkylglycerone phosphate synthase (... 171 7e-43
hsa:8540 AGPS, ADAP-S, ADAS, ADHAPS, ADPS, ALDHPSY, DKFZp762O2... 171 7e-43
dre:386801 agps, fd16e06, wu:fd16e06, zgc:56718; alkylglyceron... 171 1e-42
mmu:228061 Agps, 5832437L22, 9930035G10Rik, AW123847, Adaps, A... 170 1e-42
eco:b4463 ygcU, ECK2767, JW5442, ygcT, ygcV; predicted FAD con... 95.1 7e-20
dre:100333035 lactate dehydrogenase D-like 74.7 1e-13
ath:AT5G06580 FAD linked oxidase family protein (EC:1.1.2.4); ... 73.2 3e-13
cel:F32D8.12 hypothetical protein; K00102 D-lactate dehydrogen... 70.1 2e-12
sce:YDL174C DLD1; D-lactate dehydrogenase, oxidizes D-lactate ... 68.2 9e-12
mmu:52815 Ldhd, 4733401P21Rik, D8Bwg1320e; lactate dehydrogena... 60.8 2e-09
dre:334208 ldhd, wu:fi36b04, zgc:55447; lactate dehydrogenase ... 60.1 3e-09
sce:YDL178W DLD2, AIP2; D-lactate dehydrogenase, located in th... 58.2 1e-08
mmu:98314 D2hgdh, AA408776, AA408778, AI325464; D-2-hydroxyglu... 55.1 8e-08
hsa:197257 LDHD, DLD, MGC57726; lactate dehydrogenase D (EC:1.... 54.7 1e-07
hsa:728294 D2HGDH, D2HGD, FLJ42195, MGC25181; D-2-hydroxygluta... 54.3 1e-07
dre:565889 d2hgdh, zgc:158661; D-2-hydroxyglutarate dehydrogen... 50.8 1e-06
ath:AT4G36400 FAD linked oxidase family protein 49.3 5e-06
sce:YEL071W DLD3; D-lactate dehydrogenase, part of the retrogr... 48.5 8e-06
eco:b4468 glcE, ECK2973, gox, JW5487, yghL; glycolate oxidase ... 43.9 2e-04
eco:b2979 glcD, ECK2974, gox, JW2946, yghM; glycolate oxidase ... 41.6 0.001
cel:F54D5.12 hypothetical protein 39.7 0.003
hsa:79090 TRAPPC6A, MGC2650, TRS33; trafficking protein partic... 34.3 0.14
mmu:67091 Trappc6a, 1810073E21Rik, 4930519D19Rik, AI480686, MG... 30.8 1.4
eco:b1687 ydiJ, ECK1684, JW1677; predicted FAD-linked oxidored... 30.4 2.0
tgo:TGME49_013900 regulator of chromosome condensation, putati... 30.0 3.0
hsa:6596 HLTF, HIP116, HIP116A, HLTF1, RNF80, SMARCA3, SNF2L3,... 29.6 3.6
ath:AT2G46760 FAD-binding domain-containing protein 29.6 3.6
ath:AT4G29740 CKX4; CKX4 (CYTOKININ OXIDASE 4); amine oxidase/... 29.3 4.0
dre:565964 MGC123280, sdr42e1, si:ch211-79l17.4; zgc:123280 (E... 28.9 5.9
ath:AT1G23460 polygalacturonase 28.5 7.6
> cel:Y50D7A.7 ads-1; Alkyl-Dihydroxyacetonephosphate Synthase
family member (ads-1); K00803 alkyldihydroxyacetonephosphate
synthase [EC:2.5.1.26]
Length=597
Score = 182 bits (461), Expect = 4e-46, Method: Compositional matrix adjust.
Identities = 82/143 (57%), Positives = 108/143 (75%), Gaps = 0/143 (0%)
Query 1 VGASLEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSK 60
VG SLE +L + G T GHEPDS+EFST+GGWV+TRASGMKKN+YGNIED+VV + V K
Sbjct 212 VGQSLERQLNKKGFTCGHEPDSIEFSTLGGWVSTRASGMKKNKYGNIEDLVVHLNFVCPK 271
Query 61 GSLNQQQSAPRVSSGPSLQQLMLGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAGV 120
G + +Q PR+SSGP + Q++LGSEGTLG++++V +K+ +PE K +G FP F +GV
Sbjct 272 GIIQKQCQVPRMSSGPDIHQIILGSEGTLGVVSEVTIKIFPIPEVKRFGSFVFPNFESGV 331
Query 121 HFLRHVALDNLQPASIRLMDKTQ 143
+F R VA+ QPAS+RLMD Q
Sbjct 332 NFFREVAIQRCQPASLRLMDNDQ 354
> xla:446408 agps, MGC83829; alkylglycerone phosphate synthase
(EC:2.5.1.26); K00803 alkyldihydroxyacetonephosphate synthase
[EC:2.5.1.26]
Length=627
Score = 171 bits (433), Expect = 7e-43, Method: Compositional matrix adjust.
Identities = 81/146 (55%), Positives = 102/146 (69%), Gaps = 0/146 (0%)
Query 2 GASLEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKG 61
G LE +L G GHEPDSMEFST+GGWVATRASGMKKN YGNIED+VV + +VT KG
Sbjct 253 GQDLERQLGESGYCTGHEPDSMEFSTLGGWVATRASGMKKNIYGNIEDLVVHIKMVTPKG 312
Query 62 SLNQQQSAPRVSSGPSLQQLMLGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAGVH 121
+ + PR+S+GP + ++GSEGTLG++T+V +K++ +PE + YG + FP F GV
Sbjct 313 IIEKSCQGPRMSTGPDIHHFIMGSEGTLGVVTEVTIKIRPVPEYQKYGSVVFPNFERGVA 372
Query 122 FLRHVALDNLQPASIRLMDKTQTQCG 147
LR VA PASIRLMD Q Q G
Sbjct 373 CLREVARQRCAPASIRLMDNAQFQFG 398
> hsa:8540 AGPS, ADAP-S, ADAS, ADHAPS, ADPS, ALDHPSY, DKFZp762O2215,
FLJ99755; alkylglycerone phosphate synthase (EC:2.5.1.26);
K00803 alkyldihydroxyacetonephosphate synthase [EC:2.5.1.26]
Length=658
Score = 171 bits (433), Expect = 7e-43, Method: Compositional matrix adjust.
Identities = 79/146 (54%), Positives = 103/146 (70%), Gaps = 0/146 (0%)
Query 2 GASLEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKG 61
G LE +L+ G GHEPDS+EFSTVGGWV+TRASGMKKN YGNIED+VV + +VT +G
Sbjct 284 GQELERQLKESGYCTGHEPDSLEFSTVGGWVSTRASGMKKNIYGNIEDLVVHIKMVTPRG 343
Query 62 SLNQQQSAPRVSSGPSLQQLMLGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAGVH 121
+ + PR+S+GP + ++GSEGTLG+IT+ +K++ +PE + YG +AFP F GV
Sbjct 344 IIEKSCQGPRMSTGPDIHHFIMGSEGTLGVITEATIKIRPVPEYQKYGSVAFPNFEQGVA 403
Query 122 FLRHVALDNLQPASIRLMDKTQTQCG 147
LR +A PASIRLMD Q Q G
Sbjct 404 CLREIAKQRCAPASIRLMDNKQFQFG 429
> dre:386801 agps, fd16e06, wu:fd16e06, zgc:56718; alkylglycerone
phosphate synthase (EC:2.5.1.26); K00803 alkyldihydroxyacetonephosphate
synthase [EC:2.5.1.26]
Length=629
Score = 171 bits (432), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 79/147 (53%), Positives = 103/147 (70%), Gaps = 0/147 (0%)
Query 1 VGASLEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSK 60
+G LE +L G GHEPDSMEFS++GGWVATRASGMKKN YGNIED+VV + +VT +
Sbjct 254 IGQDLERQLNERGYCTGHEPDSMEFSSLGGWVATRASGMKKNIYGNIEDLVVHIKMVTPR 313
Query 61 GSLNQQQSAPRVSSGPSLQQLMLGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAGV 120
G + + PR+S+GP + ++GSEGTLG++T+V +K++ +PE + YG + FP F GV
Sbjct 314 GVIEKSCLGPRMSTGPDIHHFIMGSEGTLGVVTEVTMKIRPIPEYQKYGSVVFPNFQQGV 373
Query 121 HFLRHVALDNLQPASIRLMDKTQTQCG 147
LR VA PASIRLMD Q Q G
Sbjct 374 ACLREVARQRCAPASIRLMDNEQFQFG 400
> mmu:228061 Agps, 5832437L22, 9930035G10Rik, AW123847, Adaps,
Adas, Adhaps, Adps, Aldhpsy, bs2; alkylglycerone phosphate
synthase (EC:2.5.1.26); K00803 alkyldihydroxyacetonephosphate
synthase [EC:2.5.1.26]
Length=671
Score = 170 bits (430), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 78/146 (53%), Positives = 102/146 (69%), Gaps = 0/146 (0%)
Query 2 GASLEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKG 61
G LE +L+ G GHEPDS+EFSTVGGW++TRASGMKKN YGNIED+VV + +VT +G
Sbjct 297 GQDLERQLKESGYCTGHEPDSLEFSTVGGWISTRASGMKKNIYGNIEDLVVHMKMVTPRG 356
Query 62 SLNQQQSAPRVSSGPSLQQLMLGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAGVH 121
+ + PR+S+GP + ++GSEGTLG+IT+ +K++ PE + YG +AFP F GV
Sbjct 357 VIEKSSQGPRMSTGPDIHHFIMGSEGTLGVITEATIKIRPTPEYQKYGSVAFPNFEQGVA 416
Query 122 FLRHVALDNLQPASIRLMDKTQTQCG 147
LR +A PASIRLMD Q Q G
Sbjct 417 CLREIAKQRCAPASIRLMDNQQFQFG 442
> eco:b4463 ygcU, ECK2767, JW5442, ygcT, ygcV; predicted FAD containing
dehydrogenase
Length=484
Score = 95.1 bits (235), Expect = 7e-20, Method: Composition-based stats.
Identities = 49/137 (35%), Positives = 77/137 (56%), Gaps = 1/137 (0%)
Query 5 LEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGSLN 64
LE LR G T GH P S + +GG VATR+ G YG IEDMVV + V + G++
Sbjct 129 LENALREKGYTTGHSPQSKPLAQMGGLVATRSIGQFSTLYGAIEDMVVGLEAVLADGTVT 188
Query 65 QQQSAPRVSSGPSLQQLMLGSEGTLGIITQVLLKV-KLLPECKAYGCLAFPTFTAGVHFL 123
+ ++ PR ++GP ++ +++G+EG L IT+V +K+ K PE + G + L
Sbjct 189 RIKNVPRRAAGPDIRHIIIGNEGALCYITEVTVKIFKFTPENNLFYGYILEDMKTGFNIL 248
Query 124 RHVALDNLQPASIRLMD 140
R + ++ +P+ RL D
Sbjct 249 REIMVEGYRPSIARLYD 265
> dre:100333035 lactate dehydrogenase D-like
Length=484
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 47/138 (34%), Positives = 75/138 (54%), Gaps = 3/138 (2%)
Query 9 LRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGS-LNQQQ 67
LR G+ +P + +++GG ATRASG RYG ++D V+ + VVT+ G +
Sbjct 147 LRDTGLFFPIDPGAN--ASIGGMTATRASGTNAVRYGTMKDNVLAMTVVTADGEEIATGS 204
Query 68 SAPRVSSGPSLQQLMLGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAGVHFLRHVA 127
A + S+G L +L +GSEGTLG+ T + L+++ +PE G AFP+ A +
Sbjct 205 RAKKSSAGYDLTRLFVGSEGTLGVFTSITLRLQGIPEKIGGGVCAFPSVKAACDAVIMTI 264
Query 128 LDNLQPASIRLMDKTQTQ 145
+ A I L+D+ Q +
Sbjct 265 QMGIPVARIELLDEVQIR 282
> ath:AT5G06580 FAD linked oxidase family protein (EC:1.1.2.4);
K00102 D-lactate dehydrogenase (cytochrome) [EC:1.1.2.4]
Length=567
Score = 73.2 bits (178), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 44/142 (30%), Positives = 74/142 (52%), Gaps = 3/142 (2%)
Query 5 LEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGSLN 64
L E L +G+ +P +++GG ATR SG RYG + D V+ + VV G +
Sbjct 223 LNEYLEEYGLFFPLDPGPG--ASIGGMCATRCSGSLAVRYGTMRDNVISLKVVLPNGDVV 280
Query 65 QQQSAPRVSS-GPSLQQLMLGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAGVHFL 123
+ S R S+ G L +L++GSEGTLG+IT++ L+++ +P+ FPT
Sbjct 281 KTASRARKSAAGYDLTRLIIGSEGTLGVITEITLRLQKIPQHSVVAVCNFPTVKDAADVA 340
Query 124 RHVALDNLQPASIRLMDKTQTQ 145
+ +Q + + L+D+ Q +
Sbjct 341 IATMMSGIQVSRVELLDEVQIR 362
> cel:F32D8.12 hypothetical protein; K00102 D-lactate dehydrogenase
(cytochrome) [EC:1.1.2.4]
Length=460
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 49/146 (33%), Positives = 78/146 (53%), Gaps = 6/146 (4%)
Query 4 SLEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGSL 63
+L + ++ G+ +P + ++V G VAT ASG RYG +++ VV++ VV + G++
Sbjct 114 ALNDAIKNSGLFFPVDPGAD--ASVCGMVATSASGTNAIRYGTMKENVVNLEVVLADGTI 171
Query 64 NQQQ---SAPRVSS-GPSLQQLMLGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAG 119
+ PR SS G + +L +GSEGTLGIIT+ +KV P+ + +FPT
Sbjct 172 IDTKGKGRCPRKSSAGFNFTELFVGSEGTLGIITEATVKVHPRPQFLSAAVCSFPTVHEA 231
Query 120 VHFLRHVALDNLQPASIRLMDKTQTQ 145
+ V N+ A I L+D Q Q
Sbjct 232 ASTVVEVLQWNIPVARIELLDTVQIQ 257
> sce:YDL174C DLD1; D-lactate dehydrogenase, oxidizes D-lactate
to pyruvate, transcription is heme-dependent, repressed by
glucose, and derepressed in ethanol or lactate; located in
the mitochondrial inner membrane (EC:1.1.2.4); K00102 D-lactate
dehydrogenase (cytochrome) [EC:1.1.2.4]
Length=587
Score = 68.2 bits (165), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 38/140 (27%), Positives = 73/140 (52%), Gaps = 3/140 (2%)
Query 4 SLEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGSL 63
L + L HG+ G +P + +GG +A SG RYG +++ ++++ +V G++
Sbjct 230 DLNDYLSDHGLMFGCDPGPG--AQIGGCIANSCSGTNAYRYGTMKENIINMTIVLPDGTI 287
Query 64 NQQQSAPRVSS-GPSLQQLMLGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAGVHF 122
+ + PR SS G +L L +GSEGTLGI+T+ +K + P+ + ++F T
Sbjct 288 VKTKKRPRKSSAGYNLNGLFVGSEGTLGIVTEATVKCHVKPKAETVAVVSFDTIKDAAAC 347
Query 123 LRHVALDNLQPASIRLMDKT 142
++ + ++ L+D+
Sbjct 348 ASNLTQSGIHLNAMELLDEN 367
> mmu:52815 Ldhd, 4733401P21Rik, D8Bwg1320e; lactate dehydrogenase
D (EC:1.1.2.4); K00102 D-lactate dehydrogenase (cytochrome)
[EC:1.1.2.4]
Length=484
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 44/141 (31%), Positives = 68/141 (48%), Gaps = 6/141 (4%)
Query 4 SLEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGSL 63
+L LR G+ +P + +++ G AT ASG RYG + D V+++ VV G L
Sbjct 142 ALNTHLRDSGLWFPVDPGAD--ASLCGMAATGASGTNAVRYGTMRDNVINLEVVLPDGRL 199
Query 64 ----NQQQSAPRVSSGPSLQQLMLGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAG 119
+ + + ++G +L L +GSEGTLGIIT L++ PE AFP+ A
Sbjct 200 LHTAGRGRHYRKSAAGYNLTGLFVGSEGTLGIITSTTLRLHPAPEATVAATCAFPSVQAA 259
Query 120 VHFLRHVALDNLQPASIRLMD 140
V + + A I +D
Sbjct 260 VDSTVQILQAAVPVARIEFLD 280
> dre:334208 ldhd, wu:fi36b04, zgc:55447; lactate dehydrogenase
D (EC:1.1.2.4); K00102 D-lactate dehydrogenase (cytochrome)
[EC:1.1.2.4]
Length=497
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 41/141 (29%), Positives = 72/141 (51%), Gaps = 6/141 (4%)
Query 4 SLEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGSL 63
SL LR G+ +P + +++ G AT ASG RYG + + V+++ VV + G++
Sbjct 146 SLNSYLRDTGLWFPVDPGAD--ASLCGMAATSASGTNAVRYGTMRENVLNLEVVLADGTI 203
Query 64 ----NQQQSAPRVSSGPSLQQLMLGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAG 119
+ + + ++G +L L +GSEGTLGIIT+ L++ +PE +FP+ +
Sbjct 204 LHTAGKGRRPRKTAAGYNLTNLFVGSEGTLGIITKATLRLYGVPESMVSAVCSFPSVQSA 263
Query 120 VHFLRHVALDNLQPASIRLMD 140
V + + A I +D
Sbjct 264 VDSTVQILQAGVPIARIEFLD 284
> sce:YDL178W DLD2, AIP2; D-lactate dehydrogenase, located in
the mitochondrial matrix (EC:1.1.2.4)
Length=530
Score = 58.2 bits (139), Expect = 1e-08, Method: Composition-based stats.
Identities = 34/82 (41%), Positives = 51/82 (62%), Gaps = 3/82 (3%)
Query 28 VGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGSL-NQQQSAPRVSSGPSLQQLMLGSE 86
VGG VAT A G++ RYG++ V+ + VV G + N S + ++G L+QL +GSE
Sbjct 202 VGGVVATNAGGLRLLRYGSLHGSVLGLEVVMPNGQIVNSMHSMRKDNTGYDLKQLFIGSE 261
Query 87 GTLGIITQVLLKVKLLPECKAY 108
GT+GIIT V + +P+ KA+
Sbjct 262 GTIGIITGV--SILTVPKPKAF 281
> mmu:98314 D2hgdh, AA408776, AA408778, AI325464; D-2-hydroxyglutarate
dehydrogenase (EC:1.1.99.-)
Length=535
Score = 55.1 bits (131), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 35/98 (35%), Positives = 54/98 (55%), Gaps = 1/98 (1%)
Query 28 VGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGS-LNQQQSAPRVSSGPSLQQLMLGSE 86
+GG VAT A G++ RYG++ V+ + VV + G+ LN S + ++G L+Q+ +GSE
Sbjct 214 IGGNVATNAGGLRFLRYGSLRGTVLGLEVVLADGTILNCLTSLRKDNTGYDLKQMFIGSE 273
Query 87 GTLGIITQVLLKVKLLPECKAYGCLAFPTFTAGVHFLR 124
GTLG+IT V + P+ L P F + R
Sbjct 274 GTLGVITAVSIVCPPRPKAVNVAFLGCPGFAEVLQTFR 311
> hsa:197257 LDHD, DLD, MGC57726; lactate dehydrogenase D (EC:1.1.2.4);
K00102 D-lactate dehydrogenase (cytochrome) [EC:1.1.2.4]
Length=507
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 47/165 (28%), Positives = 71/165 (43%), Gaps = 29/165 (17%)
Query 4 SLEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGSL 63
+L LR G+ +P + +++ G AT ASG RYG + D V+++ VV G L
Sbjct 142 ALNAHLRDSGLWFPVDPGAD--ASLCGMAATGASGTNAVRYGTMRDNVLNLEVVLPDGRL 199
Query 64 --------------------NQQQSAPRVS-------SGPSLQQLMLGSEGTLGIITQVL 96
+ +P VS +G +L L +GSEGTLG+IT
Sbjct 200 LHTAGRGRHFRFGFWPEIPHHTAWYSPCVSLGRRKSAAGYNLTGLFVGSEGTLGLITATT 259
Query 97 LKVKLLPECKAYGCLAFPTFTAGVHFLRHVALDNLQPASIRLMDK 141
L++ PE AFP+ A V H+ + A I +D+
Sbjct 260 LRLHPAPEATVAATCAFPSVQAAVDSTVHILQAAVPVARIEFLDE 304
> hsa:728294 D2HGDH, D2HGD, FLJ42195, MGC25181; D-2-hydroxyglutarate
dehydrogenase (EC:1.1.99.-)
Length=521
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 35/91 (38%), Positives = 51/91 (56%), Gaps = 1/91 (1%)
Query 28 VGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGS-LNQQQSAPRVSSGPSLQQLMLGSE 86
+GG VAT A G++ RYG++ V+ + VV + G+ L+ S + ++G L+QL +GSE
Sbjct 200 IGGNVATNAGGLRFLRYGSLHGTVLGLEVVLADGTVLDCLTSLRKDNTGYDLKQLFIGSE 259
Query 87 GTLGIITQVLLKVKLLPECKAYGCLAFPTFT 117
GTLGIIT V + P L P F
Sbjct 260 GTLGIITTVSILCPPKPRAVNVAFLGCPGFA 290
> dre:565889 d2hgdh, zgc:158661; D-2-hydroxyglutarate dehydrogenase
(EC:1.1.99.-)
Length=533
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/69 (42%), Positives = 45/69 (65%), Gaps = 1/69 (1%)
Query 28 VGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGS-LNQQQSAPRVSSGPSLQQLMLGSE 86
+GG V+T A G++ RYG++ V+ + VV + G LN + + ++G L+QL +GSE
Sbjct 211 IGGNVSTNAGGLRLLRYGSLRGTVLGLEVVLADGHVLNCLATLRKDNTGYDLKQLFIGSE 270
Query 87 GTLGIITQV 95
GTLG+IT V
Sbjct 271 GTLGVITAV 279
> ath:AT4G36400 FAD linked oxidase family protein
Length=559
Score = 49.3 bits (116), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 46/69 (66%), Gaps = 1/69 (1%)
Query 28 VGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGS-LNQQQSAPRVSSGPSLQQLMLGSE 86
+GG V+T A G++ RYG++ V+ + VT+ G+ L+ + + ++G L+ L +GSE
Sbjct 234 IGGNVSTNAGGLRLIRYGSLHGTVLGLEAVTANGNVLDMLGTLRKDNTGYDLKHLFIGSE 293
Query 87 GTLGIITQV 95
G+LGI+T+V
Sbjct 294 GSLGIVTKV 302
> sce:YEL071W DLD3; D-lactate dehydrogenase, part of the retrograde
regulon which consists of genes whose expression is stimulated
by damage to mitochondria and reduced in cells grown
with glutamate as the sole nitrogen source, located in the
cytoplasm (EC:1.1.2.4)
Length=496
Score = 48.5 bits (114), Expect = 8e-06, Method: Composition-based stats.
Identities = 28/78 (35%), Positives = 46/78 (58%), Gaps = 1/78 (1%)
Query 28 VGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGSLNQQQSAPRV-SSGPSLQQLMLGSE 86
VGG V+T A G+ RYG++ V+ + VV G + +A R ++G L+QL +G+E
Sbjct 168 VGGVVSTNAGGLNFLRYGSLHGNVLGLEVVLPNGEIISNINALRKDNTGYDLKQLFIGAE 227
Query 87 GTLGIITQVLLKVKLLPE 104
GT+G++T V + P+
Sbjct 228 GTIGVVTGVSIVAAAKPK 245
> eco:b4468 glcE, ECK2973, gox, JW5487, yghL; glycolate oxidase
FAD binding subunit; K11472 glycolate oxidase FAD binding
subunit
Length=350
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 34/106 (32%), Positives = 55/106 (51%), Gaps = 4/106 (3%)
Query 4 SLEERLRRHGVTLGHEPDSM-EFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGS 62
++E L G L EP E +T GG VA +G ++ G++ D V+ ++T G
Sbjct 73 TIEAALESAGQMLPCEPPHYGEEATWGGMVACGLAGPRRPWSGSVRDFVLGTRIITGAGK 132
Query 63 -LNQQQSAPRVSSGPSLQQLMLGSEGTLGIITQVLLKVKLLPECKA 107
L + +G L +LM+GS G LG++T++ +KV LP +A
Sbjct 133 HLRFGGEVMKNVAGYDLSRLMVGSYGCLGVLTEISMKV--LPRPRA 176
> eco:b2979 glcD, ECK2974, gox, JW2946, yghM; glycolate oxidase
subunit, FAD-linked; K00104 glycolate oxidase [EC:1.1.3.15]
Length=499
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 44/93 (47%), Gaps = 2/93 (2%)
Query 12 HGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGSLNQQQSAPR 71
H + +P S ++GG VA A G+ +YG ++ + V T G S
Sbjct 140 HNLYYAPDPSSQIACSIGGNVAENAGGVHCLKYGLTVHNLLKIEVQTLDGEALTLGSDAL 199
Query 72 VSSGPSLQQLMLGSEGTLGIITQVLLKVKLLPE 104
S G L L GSEG LG+ T+V VKLLP+
Sbjct 200 DSPGFDLLALFTGSEGMLGVTTEV--TVKLLPK 230
> cel:F54D5.12 hypothetical protein
Length=487
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 37/148 (25%), Positives = 66/148 (44%), Gaps = 6/148 (4%)
Query 4 SLEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGS- 62
L+ +L + G + + + +GG +AT A G++ RYG++ ++ + VV
Sbjct 143 DLDNKLAKLGYMMPFDLGAKGSCQIGGNIATCAGGIRLIRYGSLHAHLLGLTVVLPDEHG 202
Query 63 --LNQQQSAPRVSSGPSLQQLMLGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAGV 120
L+ S + ++ L LGSEG LG+IT V + P+ L +F
Sbjct 203 TVLHLGSSIRKDNTTLHTPHLFLGSEGQLGVITSVTMTAVPKPKSVQSAMLGIESFKKCC 262
Query 121 HFLRHVALDNLQP--ASIRLMDKTQTQC 146
L+ +A +L +S L+D +C
Sbjct 263 EVLK-LAKSSLTEILSSFELLDDATMEC 289
> hsa:79090 TRAPPC6A, MGC2650, TRS33; trafficking protein particle
complex 6A
Length=173
Score = 34.3 bits (77), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 32/111 (28%), Positives = 51/111 (45%), Gaps = 8/111 (7%)
Query 1 VGASLEERLRRHGVTLGHEPDSMEFSTVGGWVAT---RASGMKKNRYGN--IEDMVVDVV 55
VG +L ERL R + E D ++F WVA + ++ N G ++D ++
Sbjct 58 VGQALGERLPRETLAFREELDVLKFLCKDLWVAVFQKQMDSLRTNHQGTYVLQDNSFPLL 117
Query 56 VVTSKGSLNQQQSAPRVSSGPSLQQLMLGSEGTLGIITQVLLKVKLLPECK 106
+ + G L + AP+ + L+ G+ TLGI + V V LP CK
Sbjct 118 LPMASG-LQYLEEAPKFLAFTC--GLLRGALYTLGIESVVTASVAALPVCK 165
> mmu:67091 Trappc6a, 1810073E21Rik, 4930519D19Rik, AI480686,
MGC117907, TRS33, mhyp; trafficking protein particle complex
6A
Length=159
Score = 30.8 bits (68), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 30/111 (27%), Positives = 46/111 (41%), Gaps = 8/111 (7%)
Query 1 VGASLEERLRRHGVTLGHEPDSMEFSTVGGWVAT---RASGMKKNRYGN--IEDMVVDVV 55
VG +L ERL E D+++F W A G++ N G ++D ++
Sbjct 44 VGQALGERLPLETPAFREELDALKFLCRDLWAAMFQKHMDGLRTNHQGTYVLQDNSFPLL 103
Query 56 VVTSKGSLNQQQSAPRVSSGPSLQQLMLGSEGTLGIITQVLLKVKLLPECK 106
V G + AP+ + L+ G+ TLG + V V LP CK
Sbjct 104 VTMGSGP-QYLEEAPKFLAFTC--GLLCGALHTLGFQSLVTASVASLPACK 151
> eco:b1687 ydiJ, ECK1684, JW1677; predicted FAD-linked oxidoreductase;
K06911
Length=1018
Score = 30.4 bits (67), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 13/44 (29%), Positives = 25/44 (56%), Gaps = 0/44 (0%)
Query 77 SLQQLMLGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAGV 120
L +++ GSEGTL IT+ L + LP+ + + + +F + +
Sbjct 256 DLTRILTGSEGTLAFITEARLDITRLPKVRRLVNVKYDSFDSAL 299
> tgo:TGME49_013900 regulator of chromosome condensation, putative
(EC:3.4.21.72)
Length=1155
Score = 30.0 bits (66), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 21/45 (46%), Gaps = 4/45 (8%)
Query 107 AYGCLAFPTFTAGV----HFLRHVALDNLQPASIRLMDKTQTQCG 147
AYG L FP F AG F+ H AL L + + +Q CG
Sbjct 466 AYGALGFPDFDAGAGVAPTFISHRALPALVFSGLNRPSISQVACG 510
> hsa:6596 HLTF, HIP116, HIP116A, HLTF1, RNF80, SMARCA3, SNF2L3,
ZBU1; helicase-like transcription factor
Length=1009
Score = 29.6 bits (65), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 11/33 (33%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 4 SLEERLRRHGVTLGHEPDSMEFSTVGGWVATRA 36
++ ++L++HG LG P ++ F+ GW + RA
Sbjct 159 AVSDQLKKHGFKLGPAPKTLGFNLESGWGSGRA 191
> ath:AT2G46760 FAD-binding domain-containing protein
Length=603
Score = 29.6 bits (65), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 27/78 (34%), Positives = 41/78 (52%), Gaps = 5/78 (6%)
Query 27 TVGGWVATRASGMKKNRYGN-IEDMVVDVVVVTSKGSLNQQQSAPRV---SSGPSLQQLM 82
TVGG + T A G G+ + D V ++ +V S GS+N + RV ++ P
Sbjct 168 TVGGMMGTGAHGSSLWGKGSAVHDYVTEIRIV-SPGSVNDGFAKVRVLRETTTPKEFNAA 226
Query 83 LGSEGTLGIITQVLLKVK 100
S G LG+I+QV LK++
Sbjct 227 KVSLGVLGVISQVTLKLQ 244
> ath:AT4G29740 CKX4; CKX4 (CYTOKININ OXIDASE 4); amine oxidase/
cytokinin dehydrogenase (EC:1.5.99.12); K00279 cytokinin
dehydrogenase [EC:1.5.99.12]
Length=524
Score = 29.3 bits (64), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 24/94 (25%), Positives = 46/94 (48%), Gaps = 4/94 (4%)
Query 25 FSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGSLNQQQSAPRVSSGPSLQQLMLG 84
+ +VGG ++ G + R+G V ++ V+T KG + +P+++ P L +LG
Sbjct 176 YLSVGGTLSNAGIGGQTFRHGPQISNVHELDVITGKGEM--MTCSPKLN--PELFYGVLG 231
Query 85 SEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTA 118
G GIIT+ + + P + + + F+A
Sbjct 232 GLGQFGIITRARIALDHAPTRVKWSRILYSDFSA 265
> dre:565964 MGC123280, sdr42e1, si:ch211-79l17.4; zgc:123280
(EC:1.1.1.-)
Length=402
Score = 28.9 bits (63), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 13/45 (28%), Positives = 23/45 (51%), Gaps = 1/45 (2%)
Query 9 LRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVD 53
+R+ LG+EP + V W R G K++R +I +++D
Sbjct 339 MRKAQEELGYEPKLYDLEDVVQWFQARGHGKKRSR-SSIRKLILD 382
> ath:AT1G23460 polygalacturonase
Length=460
Score = 28.5 bits (62), Expect = 7.6, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 26/50 (52%), Gaps = 2/50 (4%)
Query 83 LGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAGVHFLRHVALDNLQ 132
LG + T GI+TQV+L LL E L T+ G +++ + N++
Sbjct 292 LGKDNTTGIVTQVVLDTALLRETT--NGLRIKTYQGGSGYVQGIRFTNVE 339
Lambda K H
0.318 0.133 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2938175820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40