bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1540_orf1
Length=181
Score E
Sequences producing significant alignments: (Bits) Value
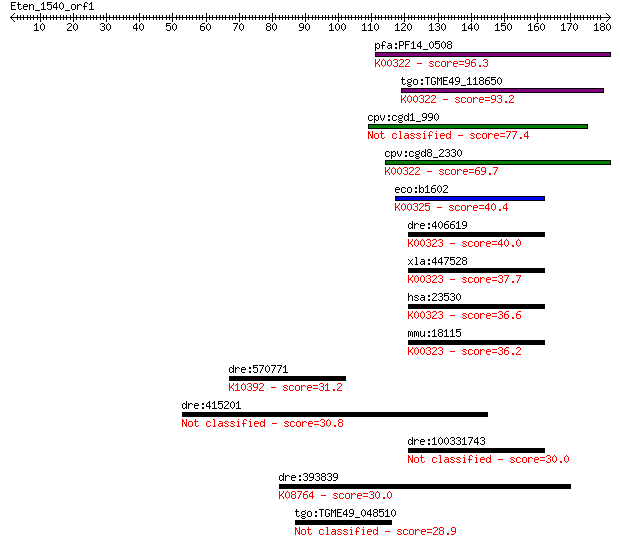
pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative (... 96.3 5e-20
tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00... 93.2 4e-19
cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alph... 77.4 3e-14
cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alp... 69.7 6e-12
eco:b1602 pntB, ECK1597, JW1594; pyridine nucleotide transhydr... 40.4 0.004
dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamid... 40.0 0.004
xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydroge... 37.7 0.022
hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide t... 36.6 0.044
mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide... 36.2 0.060
dre:570771 kif1c; kinesin family member 1C; K10392 kinesin fam... 31.2 1.8
dre:415201 zgc:86834 30.8 2.7
dre:100331743 Nicotinamide Nucleotide Transhydrogenase family ... 30.0 4.1
dre:393839 scp2, MGC77634, zgc:77634; sterol carrier protein 2... 30.0 4.6
tgo:TGME49_048510 hypothetical protein 28.9 9.5
> pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative
(EC:1.6.1.2); K00322 NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1176
Score = 96.3 bits (238), Expect = 5e-20, Method: Composition-based stats.
Identities = 42/71 (59%), Positives = 56/71 (78%), Gaps = 0/71 (0%)
Query 111 FSNMPSLLGAVYLFSAICFILCLRGLSTPQTAKRGNILGLVGMVAAVVVTFTEAGFGQHY 170
+S +PSLL AVYLFS+ICFILCL GL+ +T+KRGNILG +G+VAA+++TF++ GFG Y
Sbjct 89 YSVLPSLLNAVYLFSSICFILCLTGLNAHRTSKRGNILGFIGIVAAIMITFSQVGFGFRY 148
Query 171 LLFFATAAPAL 181
LF PA+
Sbjct 149 KLFLLIVIPAI 159
> tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00322
NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1013
Score = 93.2 bits (230), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 44/61 (72%), Positives = 50/61 (81%), Gaps = 0/61 (0%)
Query 119 GAVYLFSAICFILCLRGLSTPQTAKRGNILGLVGMVAAVVVTFTEAGFGQHYLLFFATAA 178
G VYLFSAICFIL LRGLSTP+TAKRGNILG+ GM AA+++TF+ GFG HY FF T A
Sbjct 3 GFVYLFSAICFILSLRGLSTPETAKRGNILGMTGMGAAILITFSTEGFGGHYAAFFLTVA 62
Query 179 P 179
P
Sbjct 63 P 63
> cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, 12 transmembrane domain
(EC:1.6.1.2)
Length=1147
Score = 77.4 bits (189), Expect = 3e-14, Method: Composition-based stats.
Identities = 33/66 (50%), Positives = 47/66 (71%), Gaps = 0/66 (0%)
Query 109 FAFSNMPSLLGAVYLFSAICFILCLRGLSTPQTAKRGNILGLVGMVAAVVVTFTEAGFGQ 168
+ SN SL A F+A+CFI C++GLS+ +TA R NI+G VGM+ A++V+ +E GFG
Sbjct 50 YQTSNSGSLTMAGQFFAALCFIYCIKGLSSEKTAYRSNIVGFVGMMVAILVSLSEEGFGN 109
Query 169 HYLLFF 174
HY +FF
Sbjct 110 HYFVFF 115
> cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, possible signal peptide
plus 12 transmembrane regions ; K00322 NAD(P) transhydrogenase
[EC:1.6.1.1]
Length=1143
Score = 69.7 bits (169), Expect = 6e-12, Method: Composition-based stats.
Identities = 31/68 (45%), Positives = 48/68 (70%), Gaps = 0/68 (0%)
Query 114 MPSLLGAVYLFSAICFILCLRGLSTPQTAKRGNILGLVGMVAAVVVTFTEAGFGQHYLLF 173
+ +L ++ +FS+ICF+LCLRGLST +TAKRGN LG+VG++ A+ TF F ++++F
Sbjct 87 LSTLSVSLEVFSSICFVLCLRGLSTQETAKRGNSLGIVGIICAIAATFLAPTFTMNWIMF 146
Query 174 FATAAPAL 181
A A+
Sbjct 147 IVPFALAI 154
> eco:b1602 pntB, ECK1597, JW1594; pyridine nucleotide transhydrogenase,
beta subunit (EC:1.6.1.2); K00325 NAD(P) transhydrogenase
subunit beta [EC:1.6.1.2]
Length=462
Score = 40.4 bits (93), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 19/45 (42%), Positives = 29/45 (64%), Gaps = 0/45 (0%)
Query 117 LLGAVYLFSAICFILCLRGLSTPQTAKRGNILGLVGMVAAVVVTF 161
L+ A Y+ +AI FI L GLS +T+++GN G+ GM A++ T
Sbjct 5 LVTAAYIVAAILFIFSLAGLSKHETSRQGNNFGIAGMAIALIATI 49
> dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P)
transhydrogenase [EC:1.6.1.2]
Length=1079
Score = 40.0 bits (92), Expect = 0.004, Method: Composition-based stats.
Identities = 18/41 (43%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 121 VYLFSAICFILCLRGLSTPQTAKRGNILGLVGMVAAVVVTF 161
+YL S +C + L GLST TA+ GN LG++G+ + TF
Sbjct 619 MYLGSGLCCVGALGGLSTQSTARLGNALGMIGVAGGIAATF 659
> xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydrogenase
(EC:1.6.1.2); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1086
Score = 37.7 bits (86), Expect = 0.022, Method: Composition-based stats.
Identities = 18/41 (43%), Positives = 25/41 (60%), Gaps = 0/41 (0%)
Query 121 VYLFSAICFILCLRGLSTPQTAKRGNILGLVGMVAAVVVTF 161
VYL S +C + L GLST TA+ GN LG++G+ + T
Sbjct 623 VYLGSGLCCVGALAGLSTQGTARLGNSLGMMGVAGGIAATL 663
> hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide
transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 36.6 bits (83), Expect = 0.044, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 25/41 (60%), Gaps = 0/41 (0%)
Query 121 VYLFSAICFILCLRGLSTPQTAKRGNILGLVGMVAAVVVTF 161
+YL S +C + L GLST TA+ GN LG++G+ + T
Sbjct 623 MYLGSGLCCVGALAGLSTQGTARLGNALGMIGVAGGLAATL 663
> mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 36.2 bits (82), Expect = 0.060, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 25/41 (60%), Gaps = 0/41 (0%)
Query 121 VYLFSAICFILCLRGLSTPQTAKRGNILGLVGMVAAVVVTF 161
+YL S +C + L GLST TA+ GN LG++G+ + T
Sbjct 623 MYLGSGLCCVGALGGLSTQGTARLGNALGMIGVAGGLAATL 663
> dre:570771 kif1c; kinesin family member 1C; K10392 kinesin family
member 1/13/14
Length=1180
Score = 31.2 bits (69), Expect = 1.8, Method: Composition-based stats.
Identities = 15/36 (41%), Positives = 23/36 (63%), Gaps = 1/36 (2%)
Query 67 LRSIQRHLVRMGEFFPVNRPGEG-DEGFDGARPENP 101
+RS++ +V+M + FPV+ G+ D F GAR E P
Sbjct 864 IRSLRDRMVKMEKIFPVDVKGDDEDVEFGGARVEEP 899
> dre:415201 zgc:86834
Length=177
Score = 30.8 bits (68), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 39/92 (42%), Gaps = 7/92 (7%)
Query 53 STTASAEFDFPAANLRSIQRHLVRMGEFFPVNRPGEGDEGFDGARPENPEGGAISKFAFS 112
S T A+ A L+ +Q + MG F P P + DE E+PEG A ++ + +
Sbjct 65 SNTEPADEGTTAEQLQDVQERIEAMGLFLPHPPPPDSDEE------EDPEGAA-ARRSHA 117
Query 113 NMPSLLGAVYLFSAICFILCLRGLSTPQTAKR 144
++P V L + L L P A+
Sbjct 118 SIPMDEDHVELVKRTMAAINLPTLGIPAWARE 149
> dre:100331743 Nicotinamide Nucleotide Transhydrogenase family
member (nnt-1)-like
Length=518
Score = 30.0 bits (66), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 121 VYLFSAICFILCLRGLSTPQTAKRGNILGLVGMVAAVVVTF 161
+YL S +C + L GLS T++ N LG++G+ + T
Sbjct 323 MYLGSGMCCVGALAGLSAQGTSRLRNALGMIGVAGGIAATL 363
> dre:393839 scp2, MGC77634, zgc:77634; sterol carrier protein
2 (EC:2.3.1.176); K08764 sterol carrier protein 2 [EC:2.3.1.176]
Length=538
Score = 30.0 bits (66), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 37/97 (38%), Positives = 44/97 (45%), Gaps = 18/97 (18%)
Query 82 PVNRPGE----GDEGFDGARPENPEGGAISKFAFSNMPSLLGAVYLFSAICFILC--LRG 135
P + GE GD + G NP GG ISK P LGA L A C LC LR
Sbjct 316 PEGKAGELIDRGDNTYGGKWVINPSGGLISK----GHP--LGATGL--AQCAELCWQLRA 367
Query 136 LSTPQTAKRGNILGL---VGMVAAVVVTFTEAGFGQH 169
+ P+ G L L +G+ AVVVT + GF Q
Sbjct 368 EAGPRQVP-GAKLALQHNIGLGGAVVVTLYKMGFPQE 403
> tgo:TGME49_048510 hypothetical protein
Length=11862
Score = 28.9 bits (63), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 15/29 (51%), Positives = 16/29 (55%), Gaps = 0/29 (0%)
Query 87 GEGDEGFDGARPENPEGGAISKFAFSNMP 115
GE DEG +R N EGG S F FS P
Sbjct 1140 GERDEGEGSSRQGNDEGGRASAFGFSASP 1168
Lambda K H
0.325 0.138 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4924747408
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40