bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1513_orf3
Length=230
Score E
Sequences producing significant alignments: (Bits) Value
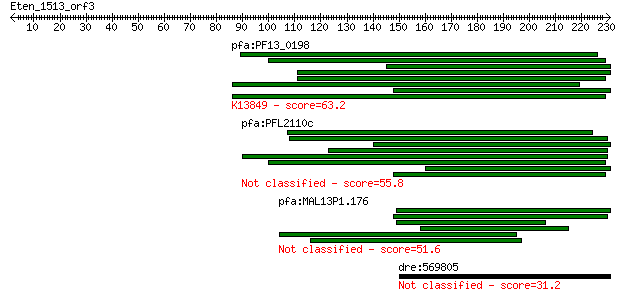
pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog A... 63.2 9e-10
pfa:PFL2110c hypothetical protein 55.8 1e-07
pfa:MAL13P1.176 PfRh2b; reticulocyte binding protein 2, homolog B 51.6 2e-06
dre:569805 im:7163673 31.2 3.1
> pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog
A; K13849 reticulocyte-binding protein
Length=3130
Score = 63.2 bits (152), Expect = 9e-10, Method: Composition-based stats.
Identities = 32/139 (23%), Positives = 83/139 (59%), Gaps = 2/139 (1%)
Query 89 RREKRTPAAAAETAAKEPQE--ASEEQQQAAEEQQQRRTVVEGAAAQAAADAAQQRYVEA 146
+R+++ E ++ QE E+Q+Q +E++ +R E + A +Q ++
Sbjct 2740 KRQEQERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQK 2799
Query 147 PEREREREQQQQQKEKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQRE 206
E + +EQ++ ++EK++Q Q+ E+ ++Q+Q ++++ + +EQ++ Q+E+E ++Q++E
Sbjct 2800 EEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELKRQEQE 2859
Query 207 KEQQQQQREKEQQQQQREK 225
+ ++++ E++Q + K
Sbjct 2860 RLERKKIELAEREQHIKSK 2878
Score = 63.2 bits (152), Expect = 9e-10, Method: Composition-based stats.
Identities = 36/131 (27%), Positives = 81/131 (61%), Gaps = 3/131 (2%)
Query 100 ETAAKEPQEASEEQQQAAEEQQQRRTVVEGAAAQAAADAAQQRYVEAPEREREREQQQQQ 159
E KE + +EQ++ E+Q++ E Q ++ ++ E+ER +++++
Sbjct 2745 ERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQER-LQKEEEL 2803
Query 160 KEKEQQQQQREKEQQQQQRE--KEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKE 217
K +EQ++ +REK++Q Q+ E K Q+Q++ +KE+ +++E+E+ Q++ E ++Q+Q+R +
Sbjct 2804 KRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELKRQEQERLER 2863
Query 218 QQQQQREKEQQ 228
++ + E+EQ
Sbjct 2864 KKIELAEREQH 2874
Score = 62.8 bits (151), Expect = 9e-10, Method: Composition-based stats.
Identities = 26/86 (30%), Positives = 66/86 (76%), Gaps = 0/86 (0%)
Query 145 EAPEREREREQQQQQKEKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQ 204
E ERE++ + Q++++ K Q+Q++ +KE+ +++E+E+ Q++ E ++Q+Q+R + ++Q+Q
Sbjct 2759 ERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELKRQEQERLEREKQEQ 2818
Query 205 REKEQQQQQREKEQQQQQREKEQQQQ 230
+KE++ +++E+E+ Q++ ++Q+Q
Sbjct 2819 LQKEEELKRQEQERLQKEEALKRQEQ 2844
Score = 61.2 bits (147), Expect = 3e-09, Method: Composition-based stats.
Identities = 35/125 (28%), Positives = 86/125 (68%), Gaps = 5/125 (4%)
Query 111 EEQQQAAEEQQQRRTVVEGAAAQAAADAAQQRYVEAPEREREREQQQQQKEKEQQQQQRE 170
E+Q+Q +E++ R+ E Q +A +++ E ++E E ++Q+Q++ + ++Q+Q +
Sbjct 2711 EKQEQLKKEEELRKKEQERQEQQQKEEALKRQEQERLQKEEELKRQEQERLEREKQEQLQ 2770
Query 171 KE-----QQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREK 225
KE Q+Q++ +KE+ +++E+E+ Q++ E ++Q+Q+R + ++Q+Q +KE++ +++E+
Sbjct 2771 KEEELKRQEQERLQKEEALKRQEQERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQ 2830
Query 226 EQQQQ 230
E+ Q+
Sbjct 2831 ERLQK 2835
Score = 60.5 bits (145), Expect = 5e-09, Method: Composition-based stats.
Identities = 35/122 (28%), Positives = 82/122 (67%), Gaps = 4/122 (3%)
Query 111 EEQQQAAEEQQQRRTVVEGAAAQAAADAAQQRYVEAPEREREREQQQQQKEKEQQQQQRE 170
+E+++ E+Q++ E + QQ+ EA +R+ + Q++++ K Q+Q++ E
Sbjct 2703 QEKERLEREKQEQLKKEEELRKKEQERQEQQQKEEALKRQEQERLQKEEELKRQEQERLE 2762
Query 171 KEQQQQ-QREKE---QQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKE 226
+E+Q+Q Q+E+E Q+Q++ +KE+ +++E+E+ Q++ E ++Q+Q+R + ++Q+Q +KE
Sbjct 2763 REKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELKRQEQERLEREKQEQLQKE 2822
Query 227 QQ 228
++
Sbjct 2823 EE 2824
Score = 56.2 bits (134), Expect = 8e-08, Method: Composition-based stats.
Identities = 29/134 (21%), Positives = 81/134 (60%), Gaps = 1/134 (0%)
Query 86 QQSRREKRTPAAAAETAAKEPQEASEEQQQAAEEQQQRRTVVEGAAAQAAADAAQQRYVE 145
++ ++E+ E +E QE +++++ ++Q+R E Q ++ ++
Sbjct 2745 ERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELK 2804
Query 146 APERER-EREQQQQQKEKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQ 204
E+ER ERE+Q+Q +++E+ ++Q ++ Q+++ K Q+Q++ +KE++ +++E+E+ +++
Sbjct 2805 RQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELKRQEQERLERK 2864
Query 205 REKEQQQQQREKEQ 218
+ + +++Q K +
Sbjct 2865 KIELAEREQHIKSK 2878
Score = 43.9 bits (102), Expect = 5e-04, Method: Composition-based stats.
Identities = 26/96 (27%), Positives = 70/96 (72%), Gaps = 13/96 (13%)
Query 148 ERER-EREQQQQQKEKEQQQQQREK-----------EQQQQQREKEQQQQQREKEQQQQQ 195
E+ER ERE+Q+Q K++E+ +++ ++ ++Q+Q+R +++++ +R+++++ ++
Sbjct 2704 EKERLEREKQEQLKKEEELRKKEQERQEQQQKEEALKRQEQERLQKEEELKRQEQERLER 2763
Query 196 REKEQQQQQREKEQQQQQR-EKEQQQQQREKEQQQQ 230
++EQ Q++ E ++Q+Q+R +KE+ +++E+E+ Q+
Sbjct 2764 EKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQK 2799
Score = 37.0 bits (84), Expect = 0.056, Method: Composition-based stats.
Identities = 27/147 (18%), Positives = 74/147 (50%), Gaps = 4/147 (2%)
Query 86 QQSRREKRTPAAAAETAAKEPQEASEEQQQAAEEQQQR--RTVVEGAAAQAAADAAQQRY 143
++ +R+++ E ++ QE +++++ ++Q+R R E + +Q
Sbjct 2773 EELKRQEQERLQKEEALKRQEQERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQER 2832
Query 144 VEAPEREREREQQQQQKEKE--QQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQ 201
++ E + +EQ++ QKE+E +Q+Q+R + ++ + E+EQ + + + + + E
Sbjct 2833 LQKEEALKRQEQERLQKEEELKRQEQERLERKKIELAEREQHIKSKLESDMVKIIKDELT 2892
Query 202 QQQREKEQQQQQREKEQQQQQREKEQQ 228
+++ E + + + + +Q+ K Q
Sbjct 2893 KEKDEIIKNKDIKLRHSLEQKWLKHLQ 2919
> pfa:PFL2110c hypothetical protein
Length=1846
Score = 55.8 bits (133), Expect = 1e-07, Method: Composition-based stats.
Identities = 27/122 (22%), Positives = 77/122 (63%), Gaps = 12/122 (9%)
Query 108 EASEEQQQAAEEQQQRRTVVEGAAAQAAADAAQQRYVEAPEREREREQQQQQKEKEQQQQ 167
E +E+Q+ EEQ+ G + E ++ RE+Q+ ++E++ +++
Sbjct 1498 ENMKEEQKMREEQKVGEEQKVGEEQKVG------------EEQKLREEQKMREEQKMREE 1545
Query 168 QREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQ 227
Q+ +E+Q+ + E++ +++Q+ +E+Q+ + E++ +++Q+ +E+Q+ + E++ +++Q+ +E+
Sbjct 1546 QKMREEQKMREEQKVREEQKMREEQKMREEQKMREEQKVREEQKLREEQKMREEQKMREE 1605
Query 228 QQ 229
Q+
Sbjct 1606 QK 1607
Score = 55.8 bits (133), Expect = 1e-07, Method: Composition-based stats.
Identities = 24/117 (20%), Positives = 72/117 (61%), Gaps = 0/117 (0%)
Query 107 QEASEEQQQAAEEQQQRRTVVEGAAAQAAADAAQQRYVEAPEREREREQQQQQKEKEQQQ 166
+E + EEQ+ R G + + + E ++ RE+Q+ ++E++ ++
Sbjct 1491 EEKMSADENMKEEQKMREEQKVGEEQKVGEEQKVGEEQKLREEQKMREEQKMREEQKMRE 1550
Query 167 QQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQR 223
+Q+ +E+Q+ + E++ +++Q+ +E+Q+ + E++ +++Q+ +E+Q+ + E++ +++Q+
Sbjct 1551 EQKMREEQKVREEQKMREEQKMREEQKMREEQKVREEQKLREEQKMREEQKMREEQK 1607
Score = 50.8 bits (120), Expect = 4e-06, Method: Composition-based stats.
Identities = 20/91 (21%), Positives = 65/91 (71%), Gaps = 0/91 (0%)
Query 140 QQRYVEAPEREREREQQQQQKEKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKE 199
+Q+ E + E++ ++QK E+Q+ + E++ +++Q+ +E+Q+ + E++ +++Q+ +E
Sbjct 1503 EQKMREEQKVGEEQKVGEEQKVGEEQKLREEQKMREEQKMREEQKMREEQKMREEQKVRE 1562
Query 200 QQQQQREKEQQQQQREKEQQQQQREKEQQQQ 230
+Q+ + E++ +++Q+ +E+Q+ + E++ +++
Sbjct 1563 EQKMREEQKMREEQKMREEQKVREEQKLREE 1593
Score = 48.1 bits (113), Expect = 3e-05, Method: Composition-based stats.
Identities = 25/109 (22%), Positives = 69/109 (63%), Gaps = 2/109 (1%)
Query 123 RRTVVEGAAAQAAADAAQQRYVEAPEREREREQQQQQKEKEQQQQQREKEQQQQQREKEQ 182
RR E +A Q+ E E ++ ++Q+ +EQ+ ++ +K +++Q+ +EQ
Sbjct 1487 RRGEEEKMSADENMKEEQKMREEQKVGEEQKVGEEQKVGEEQKLREEQKMREEQKMREEQ 1546
Query 183 Q--QQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQ 229
+ ++Q+ +E+Q+ + E++ +++Q+ +E+Q+ + E++ +++Q+ +E+Q+
Sbjct 1547 KMREEQKMREEQKVREEQKMREEQKMREEQKMREEQKVREEQKLREEQK 1595
Score = 44.7 bits (104), Expect = 3e-04, Method: Composition-based stats.
Identities = 34/152 (22%), Positives = 86/152 (56%), Gaps = 13/152 (8%)
Query 90 REKRTPAAAAETAAKEPQEASEEQQQAAEEQQQRRTVVEGAAAQAAADAAQQRYVEAPER 149
R +A+ KE Q+ EEQ+ EEQ+ G + + + + E
Sbjct 1487 RRGEEEKMSADENMKEEQKMREEQK-VGEEQKVGEEQKVGEEQKLREEQKMREEQKMREE 1545
Query 150 EREREQQQQQKEKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQ 209
++ RE+Q+ ++E++ +++Q+ +E+Q+ + E++ +++Q+ +E+Q+ + E++ +++Q+ +E+
Sbjct 1546 QKMREEQKMREEQKVREEQKMREEQKMREEQKMREEQKVREEQKLREEQKMREEQKMREE 1605
Query 210 Q------------QQQREKEQQQQQREKEQQQ 229
Q Q +REK+ ++++++K Q+
Sbjct 1606 QKMREEEKIREEEQMKREKKMRREEKKKRVQE 1637
Score = 38.9 bits (89), Expect = 0.016, Method: Composition-based stats.
Identities = 35/146 (23%), Positives = 80/146 (54%), Gaps = 20/146 (13%)
Query 100 ETAAKEPQEASEEQ-----QQAAEEQQQRRTVVEGAAAQAAADAAQQRYVEAPERERERE 154
E +E Q+ EEQ Q+ EEQ+ R E + +Q+ E + E++
Sbjct 1503 EQKMREEQKVGEEQKVGEEQKVGEEQKLRE---EQKMREEQKMREEQKMREEQKMREEQK 1559
Query 155 QQQQQKEKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQ------------Q 202
+++QK +E+Q+ + E++ +++Q+ +E+Q+ + E++ +++Q+ +E+Q Q
Sbjct 1560 VREEQKMREEQKMREEQKMREEQKVREEQKLREEQKMREEQKMREEQKMREEEKIREEEQ 1619
Query 203 QQREKEQQQQQREKEQQQQQREKEQQ 228
+REK+ ++++++K Q+ + E Q
Sbjct 1620 MKREKKMRREEKKKRVQEPIKIDEIQ 1645
Score = 35.4 bits (80), Expect = 0.18, Method: Composition-based stats.
Identities = 14/71 (19%), Positives = 49/71 (69%), Gaps = 1/71 (1%)
Query 160 KEKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQ 219
+ E+++ ++ +++Q+ +E+Q+ E++ ++Q+ E+Q+ + E++ +++Q+ +E+Q
Sbjct 1487 RRGEEEKMSADENMKEEQKMREEQKVGEEQKVGEEQKVGEEQKLREEQKMREEQKMREEQ 1546
Query 220 QQQREKEQQQQ 230
+ RE+++ ++
Sbjct 1547 KM-REEQKMRE 1556
Score = 31.2 bits (69), Expect = 2.9, Method: Composition-based stats.
Identities = 24/83 (28%), Positives = 53/83 (63%), Gaps = 5/83 (6%)
Query 148 EREREREQQQQQK-EKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREK-EQQQQQR 205
E+E+++ Q+ +K EK+++Q Q+EK +++ EKE+Q EK + Q+++E E ++ +R
Sbjct 1385 EQEKKKLQEDIKKLEKDKEQFQKEKIRRE---EKEKQLLLEEKIKLQKEKELFENEKLER 1441
Query 206 EKEQQQQQREKEQQQQQREKEQQ 228
+ + E E+++ +R K ++
Sbjct 1442 KMSYMLKINELEKKKNERNKMEK 1464
> pfa:MAL13P1.176 PfRh2b; reticulocyte binding protein 2, homolog
B
Length=3179
Score = 51.6 bits (122), Expect = 2e-06, Method: Composition-based stats.
Identities = 24/82 (29%), Positives = 69/82 (84%), Gaps = 0/82 (0%)
Query 149 REREREQQQQQKEKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKE 208
+E+ER ++++Q++ +++ +++++++Q+Q++KE+ +++E+E+ Q++ E ++Q+Q+R +
Sbjct 2628 QEKERLEREKQEQLKKEALKKQEQERQEQQQKEEALKRQEQERLQKEEELKRQEQERLER 2687
Query 209 QQQQQREKEQQQQQREKEQQQQ 230
++Q+Q +KE++ +++E+E+QQQ
Sbjct 2688 EKQEQLQKEEELRKKEQEKQQQ 2709
Score = 50.8 bits (120), Expect = 4e-06, Method: Composition-based stats.
Identities = 26/82 (31%), Positives = 65/82 (79%), Gaps = 0/82 (0%)
Query 148 EREREREQQQQQKEKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREK 207
E+ER ++Q+Q +KE ++Q ++ Q+QQQ+E+ ++Q++E+ Q++++ ++++Q++ +
Sbjct 2629 EKERLEREKQEQLKKEALKKQEQERQEQQQKEEALKRQEQERLQKEEELKRQEQERLERE 2688
Query 208 EQQQQQREKEQQQQQREKEQQQ 229
+Q+Q Q+E+E +++++EK+QQ+
Sbjct 2689 KQEQLQKEEELRKKEQEKQQQR 2710
Score = 49.7 bits (117), Expect = 9e-06, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 47/57 (82%), Gaps = 0/57 (0%)
Query 149 REREREQQQQQKEKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQR 205
+ +E+E+ Q+++E ++Q+Q+R + ++Q+Q +KE++ +++E+E+QQQ+ +E ++Q++
Sbjct 2664 KRQEQERLQKEEELKRQEQERLEREKQEQLQKEEELRKKEQEKQQQRNIQELEEQKK 2720
Score = 49.7 bits (117), Expect = 9e-06, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 47/57 (82%), Gaps = 0/57 (0%)
Query 158 QQKEKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQREKEQQQQQR 214
+++E+E+ Q++ E ++Q+Q+R + ++Q+Q +KE++ +++E+E+QQQ+ +E ++Q++
Sbjct 2664 KRQEQERLQKEEELKRQEQERLEREKQEQLQKEEELRKKEQEKQQQRNIQELEEQKK 2720
Score = 42.7 bits (99), Expect = 0.001, Method: Composition-based stats.
Identities = 23/91 (25%), Positives = 55/91 (60%), Gaps = 0/91 (0%)
Query 104 KEPQEASEEQQQAAEEQQQRRTVVEGAAAQAAADAAQQRYVEAPEREREREQQQQQKEKE 163
KE E +++Q E +++ + + A Q++ E E +R++Q++ + ++
Sbjct 2630 KERLEREKQEQLKKEALKKQEQERQEQQQKEEALKRQEQERLQKEEELKRQEQERLEREK 2689
Query 164 QQQQQREKEQQQQQREKEQQQQQREKEQQQQ 194
Q+Q Q+E+E +++++EK+QQ+ +E E+Q++
Sbjct 2690 QEQLQKEEELRKKEQEKQQQRNIQELEEQKK 2720
Score = 38.5 bits (88), Expect = 0.023, Method: Composition-based stats.
Identities = 19/81 (23%), Positives = 46/81 (56%), Gaps = 22/81 (27%)
Query 116 AAEEQQQRRTVVEGAAAQAAADAAQQRYVEAPEREREREQQQQQKEKEQQQQQREKEQQQ 175
A + Q+Q R ++E E ++Q+Q++ + ++Q+Q +KE++
Sbjct 2662 ALKRQEQERL----------------------QKEEELKRQEQERLEREKQEQLQKEEEL 2699
Query 176 QQREKEQQQQQREKEQQQQQR 196
+++E+E+QQQ+ +E ++Q++
Sbjct 2700 RKKEQEKQQQRNIQELEEQKK 2720
> dre:569805 im:7163673
Length=1913
Score = 31.2 bits (69), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 67/93 (72%), Gaps = 12/93 (12%)
Query 150 EREREQQQQQKEKEQQQQQREK-EQQQQQREKEQQQQ-QREKEQQQQQREKEQ------- 200
E ER Q+++ + KEQ+ ++ E+ E ++Q+ E+EQ++Q +RE+++++Q+ E+ Q
Sbjct 1360 ETERLQKEENERKEQEHEESERAETERQKLEQEQKEQLKREEDEKRQENERIQREKDEER 1419
Query 201 ---QQQQREKEQQQQQREKEQQQQQREKEQQQQ 230
++ ++++ +Q+ QR+KE+++ QREKEQ+++
Sbjct 1420 EEKERLEKKRNEQEDQRQKEEERIQREKEQKEK 1452
Lambda K H
0.304 0.117 0.305
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7761100204
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40