bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1498_orf1
Length=88
Score E
Sequences producing significant alignments: (Bits) Value
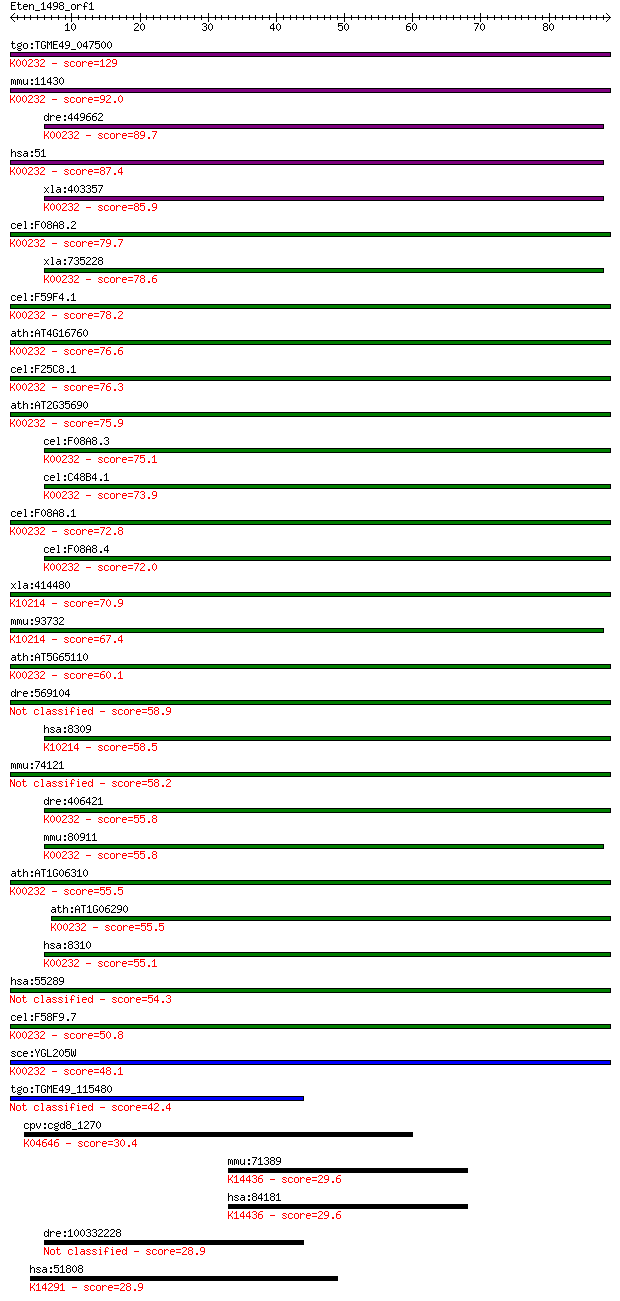
tgo:TGME49_047500 acyl-coenzyme A oxidase, putative (EC:1.9.3.... 129 2e-30
mmu:11430 Acox1, AI042784, AOX, D130055E20Rik; acyl-Coenzyme A... 92.0 5e-19
dre:449662 acox1, wu:fb59h12, zgc:114033, zgc:92584; acyl-Coen... 89.7 2e-18
hsa:51 ACOX1, ACOX, MGC1198, PALMCOX, SCOX; acyl-CoA oxidase 1... 87.4 9e-18
xla:403357 hypothetical protein MGC68531; K00232 acyl-CoA oxid... 85.9 3e-17
cel:F08A8.2 hypothetical protein; K00232 acyl-CoA oxidase [EC:... 79.7 2e-15
xla:735228 acox1, MGC131363, aco, acox; acyl-CoA oxidase 1, pa... 78.6 5e-15
cel:F59F4.1 hypothetical protein; K00232 acyl-CoA oxidase [EC:... 78.2 6e-15
ath:AT4G16760 ACX1; ACX1 (ACYL-COA OXIDASE 1); acyl-CoA oxidas... 76.6 2e-14
cel:F25C8.1 hypothetical protein; K00232 acyl-CoA oxidase [EC:... 76.3 2e-14
ath:AT2G35690 ACX5; ACX5 (ACYL-COA OXIDASE 5); FAD binding / a... 75.9 3e-14
cel:F08A8.3 hypothetical protein; K00232 acyl-CoA oxidase [EC:... 75.1 6e-14
cel:C48B4.1 hypothetical protein; K00232 acyl-CoA oxidase [EC:... 73.9 1e-13
cel:F08A8.1 hypothetical protein; K00232 acyl-CoA oxidase [EC:... 72.8 2e-13
cel:F08A8.4 hypothetical protein; K00232 acyl-CoA oxidase [EC:... 72.0 4e-13
xla:414480 acox2, MGC83074; acyl-CoA oxidase 2, branched chain... 70.9 9e-13
mmu:93732 Acox2, THCCox; acyl-Coenzyme A oxidase 2, branched c... 67.4 1e-11
ath:AT5G65110 ACX2; ACX2 (ACYL-COA OXIDASE 2); acyl-CoA oxidas... 60.1 1e-09
dre:569104 si:ch211-205h19.1 58.9 4e-09
hsa:8309 ACOX2, BCOX, BRCACOX, BRCOX, THCCox; acyl-CoA oxidase... 58.5 4e-09
mmu:74121 Acoxl, 1200014P05Rik, AV025673; acyl-Coenzyme A oxid... 58.2 6e-09
dre:406421 acox3, zgc:64087; acyl-Coenzyme A oxidase 3, prista... 55.8 3e-08
mmu:80911 Acox3, BI685180, EST-s59, PCOX; acyl-Coenzyme A oxid... 55.8 3e-08
ath:AT1G06310 ACX6; ACX6 (ACYL-COA OXIDASE 6); FAD binding / a... 55.5 4e-08
ath:AT1G06290 ACX3; ACX3 (ACYL-COA OXIDASE 3); acyl-CoA oxidas... 55.5 4e-08
hsa:8310 ACOX3; acyl-CoA oxidase 3, pristanoyl (EC:1.3.3.6); K... 55.1 5e-08
hsa:55289 ACOXL, FLJ11042; acyl-CoA oxidase-like 54.3 8e-08
cel:F58F9.7 hypothetical protein; K00232 acyl-CoA oxidase [EC:... 50.8 1e-06
sce:YGL205W POX1, FOX1; Fatty-acyl coenzyme A oxidase, involve... 48.1 7e-06
tgo:TGME49_115480 acyl-CoA oxidase, putative (EC:1.9.3.1) 42.4 3e-04
cpv:cgd8_1270 clathrin heavy chain ; K04646 clathrin heavy chain 30.4 1.3
mmu:71389 Chd6, 5430439G14Rik, 6330406J24Rik; chromodomain hel... 29.6 2.2
hsa:84181 CHD6, CHD5, KIAA1335, RIGB; chromodomain helicase DN... 29.6 2.4
dre:100332228 nucleotide-binding oligomerization domain contai... 28.9 3.7
hsa:51808 PHAX, FLJ13193, RNUXA; phosphorylated adaptor for RN... 28.9 4.2
> tgo:TGME49_047500 acyl-coenzyme A oxidase, putative (EC:1.9.3.1);
K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=696
Score = 129 bits (325), Expect = 2e-30, Method: Composition-based stats.
Identities = 56/88 (63%), Positives = 72/88 (81%), Gaps = 0/88 (0%)
Query 1 VTLLHIGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTRSRKLLYSTLTF 60
V L HIG+K+GY +DNG + L NVRIPR+NLLM++C VDR GTY+ S+KL+Y T+TF
Sbjct 233 VKLFHIGKKLGYNSMDNGGVQLHNVRIPRRNLLMKYCHVDRQGTYKPLGSKKLMYGTMTF 292
Query 61 TRKQIIRSAGAHLSRSIVVAIRYSAVRR 88
TRKQII +AG L++S+V+AIRYSAVRR
Sbjct 293 TRKQIIMAAGPQLAKSVVIAIRYSAVRR 320
> mmu:11430 Acox1, AI042784, AOX, D130055E20Rik; acyl-Coenzyme
A oxidase 1, palmitoyl (EC:1.3.3.6); K00232 acyl-CoA oxidase
[EC:1.3.3.6]
Length=661
Score = 92.0 bits (227), Expect = 5e-19, Method: Composition-based stats.
Identities = 43/88 (48%), Positives = 59/88 (67%), Gaps = 0/88 (0%)
Query 1 VTLLHIGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTRSRKLLYSTLTF 60
+T+ IG K GY+ +DNG L + N RIPR+N+LM++ +V DGTY S KL Y T+ F
Sbjct 221 ITVGDIGPKFGYEEMDNGYLKMDNYRIPRENMLMKYAQVKPDGTYVKPLSNKLTYGTMVF 280
Query 61 TRKQIIRSAGAHLSRSIVVAIRYSAVRR 88
R ++ SA LS++ +AIRYSAVRR
Sbjct 281 VRSFLVGSAAQSLSKACTIAIRYSAVRR 308
> dre:449662 acox1, wu:fb59h12, zgc:114033, zgc:92584; acyl-Coenzyme
A oxidase 1, palmitoyl (EC:1.3.3.6); K00232 acyl-CoA
oxidase [EC:1.3.3.6]
Length=660
Score = 89.7 bits (221), Expect = 2e-18, Method: Composition-based stats.
Identities = 43/82 (52%), Positives = 56/82 (68%), Gaps = 0/82 (0%)
Query 6 IGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTRSRKLLYSTLTFTRKQI 65
IG K G+ VDNG L L+NVRIPR+N+LM++ +V+ DGTY S KL Y T+ F R I
Sbjct 226 IGPKFGFDEVDNGYLKLENVRIPRENMLMKYAQVEPDGTYVKPPSDKLTYGTMVFIRSMI 285
Query 66 IRSAGAHLSRSIVVAIRYSAVR 87
+ + LS+S +AIRYSAVR
Sbjct 286 VGESARALSKSCTIAIRYSAVR 307
> hsa:51 ACOX1, ACOX, MGC1198, PALMCOX, SCOX; acyl-CoA oxidase
1, palmitoyl (EC:1.3.3.6); K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=622
Score = 87.4 bits (215), Expect = 9e-18, Method: Composition-based stats.
Identities = 41/87 (47%), Positives = 56/87 (64%), Gaps = 0/87 (0%)
Query 1 VTLLHIGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTRSRKLLYSTLTF 60
+T+ IG K GY +DNG L + N RIPR+N+LM++ +V DGTY S KL Y T+ F
Sbjct 183 ITVGDIGPKFGYDEIDNGYLKMDNHRIPRENMLMKYAQVKPDGTYVKPLSNKLTYGTMVF 242
Query 61 TRKQIIRSAGAHLSRSIVVAIRYSAVR 87
R ++ A LS++ +AIRYSAVR
Sbjct 243 VRSFLVGEAARALSKACTIAIRYSAVR 269
> xla:403357 hypothetical protein MGC68531; K00232 acyl-CoA oxidase
[EC:1.3.3.6]
Length=661
Score = 85.9 bits (211), Expect = 3e-17, Method: Composition-based stats.
Identities = 41/82 (50%), Positives = 52/82 (63%), Gaps = 0/82 (0%)
Query 6 IGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTRSRKLLYSTLTFTRKQI 65
IG K GY DNG L VRIPR+N+LM++ +V+ DGTY S KL Y T+ F R I
Sbjct 226 IGPKFGYDETDNGFLKFDKVRIPRENMLMKYAKVEPDGTYVKPLSDKLTYGTMVFIRSMI 285
Query 66 IRSAGAHLSRSIVVAIRYSAVR 87
+ + LSR+ +AIRYSAVR
Sbjct 286 VGDSARSLSRACTIAIRYSAVR 307
> cel:F08A8.2 hypothetical protein; K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=679
Score = 79.7 bits (195), Expect = 2e-15, Method: Composition-based stats.
Identities = 40/88 (45%), Positives = 54/88 (61%), Gaps = 0/88 (0%)
Query 1 VTLLHIGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTRSRKLLYSTLTF 60
VT+ IG K+ + DNG L L N+R+PR NLLMR C+V+ DGTY K+ YS +
Sbjct 232 VTVGDIGPKMNFNAADNGYLGLNNLRVPRTNLLMRHCKVEADGTYVKPPHAKIGYSGMVK 291
Query 61 TRKQIIRSAGAHLSRSIVVAIRYSAVRR 88
R Q+ G L+ ++ +A RYSAVRR
Sbjct 292 IRSQMAMEQGLFLAHALTIAARYSAVRR 319
> xla:735228 acox1, MGC131363, aco, acox; acyl-CoA oxidase 1,
palmitoyl (EC:1.3.3.6); K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=661
Score = 78.6 bits (192), Expect = 5e-15, Method: Composition-based stats.
Identities = 36/82 (43%), Positives = 50/82 (60%), Gaps = 0/82 (0%)
Query 6 IGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTRSRKLLYSTLTFTRKQI 65
IG K G+ DNG L VRIPR+ ++M++ +V+ DGTY S KL Y T+ F R I
Sbjct 226 IGPKFGFDETDNGFLKFDKVRIPREYMMMKYAKVEPDGTYVKPLSDKLTYGTMVFIRSMI 285
Query 66 IRSAGAHLSRSIVVAIRYSAVR 87
+ + LSR+ +A+RYS VR
Sbjct 286 VGDSARSLSRACTIAVRYSTVR 307
> cel:F59F4.1 hypothetical protein; K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=667
Score = 78.2 bits (191), Expect = 6e-15, Method: Composition-based stats.
Identities = 40/88 (45%), Positives = 52/88 (59%), Gaps = 0/88 (0%)
Query 1 VTLLHIGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTRSRKLLYSTLTF 60
+ L IG K+G G DNG L VRIPRK LLMR+ +V+ DGTY KL Y T+ F
Sbjct 231 IRLGDIGPKLGINGNDNGFLLFDKVRIPRKALLMRYAKVNPDGTYIAPAHSKLGYGTMVF 290
Query 61 TRKQIIRSAGAHLSRSIVVAIRYSAVRR 88
R +I+ L+ + +A RY+AVRR
Sbjct 291 VRSIMIKDQSTQLAAAATIATRYAAVRR 318
> ath:AT4G16760 ACX1; ACX1 (ACYL-COA OXIDASE 1); acyl-CoA oxidase
(EC:1.3.3.6); K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=664
Score = 76.6 bits (187), Expect = 2e-14, Method: Composition-based stats.
Identities = 39/92 (42%), Positives = 58/92 (63%), Gaps = 4/92 (4%)
Query 1 VTLLHIGQKIG---YQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTR-SRKLLYS 56
+T+ IG K+G Y +DNG L +VRIPR +LMR +V R+G Y + ++L+Y
Sbjct 220 ITVGDIGTKMGNGAYNSMDNGFLMFDHVRIPRDQMLMRLSKVTREGEYVPSDVPKQLVYG 279
Query 57 TLTFTRKQIIRSAGAHLSRSIVVAIRYSAVRR 88
T+ + R+ I+ A LSR++ +A RYSAVRR
Sbjct 280 TMVYVRQTIVADASNALSRAVCIATRYSAVRR 311
> cel:F25C8.1 hypothetical protein; K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=601
Score = 76.3 bits (186), Expect = 2e-14, Method: Composition-based stats.
Identities = 37/88 (42%), Positives = 50/88 (56%), Gaps = 0/88 (0%)
Query 1 VTLLHIGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTRSRKLLYSTLTF 60
+ L IG K+G G DNG L N RI R N+LMR +V DGTY+ KL Y + F
Sbjct 230 IKLGDIGPKLGSNGSDNGYLVFTNYRISRGNMLMRHSKVHPDGTYQKPPHSKLAYGGMVF 289
Query 61 TRKQIIRSAGAHLSRSIVVAIRYSAVRR 88
R ++R +L+ ++ +A RYS VRR
Sbjct 290 VRSMMVRDIANYLANAVTIATRYSTVRR 317
> ath:AT2G35690 ACX5; ACX5 (ACYL-COA OXIDASE 5); FAD binding /
acyl-CoA dehydrogenase/ acyl-CoA oxidase/ electron carrier/
oxidoreductase/ oxidoreductase, acting on the CH-CH group of
donors (EC:1.3.3.6); K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=664
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 38/92 (41%), Positives = 57/92 (61%), Gaps = 4/92 (4%)
Query 1 VTLLHIGQKIG---YQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTR-SRKLLYS 56
+T+ IG K G Y +DNG L + RIPR +LMR +V R+G Y + R+L+Y
Sbjct 220 ITVGDIGMKFGNGAYNSMDNGFLMFDHFRIPRDQMLMRLSKVTREGKYVASDVPRQLVYG 279
Query 57 TLTFTRKQIIRSAGAHLSRSIVVAIRYSAVRR 88
T+ + R+ I+ +A L+R++ +A RYSAVRR
Sbjct 280 TMVYVRQSIVSNASTALARAVCIATRYSAVRR 311
> cel:F08A8.3 hypothetical protein; K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=660
Score = 75.1 bits (183), Expect = 6e-14, Method: Composition-based stats.
Identities = 37/83 (44%), Positives = 52/83 (62%), Gaps = 0/83 (0%)
Query 6 IGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTRSRKLLYSTLTFTRKQI 65
IG K+ + G DNG L N RIPR NLLMR +V+ +GTY K+ YS++ R ++
Sbjct 236 IGPKMAFNGADNGYLGFNNHRIPRTNLLMRHTKVEANGTYIKPSHAKIGYSSMVKVRSRM 295
Query 66 IRSAGAHLSRSIVVAIRYSAVRR 88
G L+ ++V+A+RYSAVRR
Sbjct 296 AMDQGLFLASALVIAVRYSAVRR 318
> cel:C48B4.1 hypothetical protein; K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=659
Score = 73.9 bits (180), Expect = 1e-13, Method: Composition-based stats.
Identities = 34/83 (40%), Positives = 53/83 (63%), Gaps = 0/83 (0%)
Query 6 IGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTRSRKLLYSTLTFTRKQI 65
IG K+G VDNG L N RIPR+N+LM+ +V ++G Y K+ Y+T+ + R ++
Sbjct 238 IGTKMGVNCVDNGFLAFDNYRIPRRNMLMKHSKVSKEGLYTAPSHPKVGYTTMLYMRSEM 297
Query 66 IRSAGAHLSRSIVVAIRYSAVRR 88
I +L+ ++ ++IRYSAVRR
Sbjct 298 IYHQAYYLAMAMAISIRYSAVRR 320
> cel:F08A8.1 hypothetical protein; K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=674
Score = 72.8 bits (177), Expect = 2e-13, Method: Composition-based stats.
Identities = 39/88 (44%), Positives = 51/88 (57%), Gaps = 0/88 (0%)
Query 1 VTLLHIGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTRSRKLLYSTLTF 60
+T+ IG K+ Y VDNG L N RIPR NLLMR +V+ DGTY K+ YS +
Sbjct 234 ITIGDIGPKMAYNIVDNGFLGFNNYRIPRTNLLMRHTKVEADGTYIKPPHAKINYSAMVH 293
Query 61 TRKQIIRSAGAHLSRSIVVAIRYSAVRR 88
R ++ LS ++ +A RYSAVRR
Sbjct 294 VRSYMLTGQAIMLSYALNIATRYSAVRR 321
> cel:F08A8.4 hypothetical protein; K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=662
Score = 72.0 bits (175), Expect = 4e-13, Method: Composition-based stats.
Identities = 35/83 (42%), Positives = 50/83 (60%), Gaps = 0/83 (0%)
Query 6 IGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTRSRKLLYSTLTFTRKQI 65
IG K+ + G DNG L R+PR NLLMR V+ DGTY K+ +S + R +
Sbjct 238 IGPKMSFNGGDNGFLGFDKFRVPRTNLLMRHVRVEADGTYVKPPHAKVNHSAMVHVRSHM 297
Query 66 IRSAGAHLSRSIVVAIRYSAVRR 88
GA L++++++A+RYSAVRR
Sbjct 298 ATGQGALLAQALIIAVRYSAVRR 320
> xla:414480 acox2, MGC83074; acyl-CoA oxidase 2, branched chain;
K10214 3alpha,7alpha,12alpha-trihydroxy-5beta-cholestanoyl-CoA
24-hydroxylase [EC:1.17.99.3]
Length=670
Score = 70.9 bits (172), Expect = 9e-13, Method: Composition-based stats.
Identities = 35/89 (39%), Positives = 57/89 (64%), Gaps = 1/89 (1%)
Query 1 VTLLHIGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTRSRKLLYSTLTF 60
+T+ IG K+ ++ +DNG L ++N+R+P++N+L RF EV DGTY S K+ Y T+
Sbjct 231 ITVGDIGPKMSFEHIDNGFLMMRNIRVPKENMLSRFSEVLPDGTYVKRGSDKINYFTMVA 290
Query 61 TRKQIIRSAGAH-LSRSIVVAIRYSAVRR 88
R ++R L ++ ++IRY+AVRR
Sbjct 291 VRVSMLRQEVLEALMKACTISIRYAAVRR 319
> mmu:93732 Acox2, THCCox; acyl-Coenzyme A oxidase 2, branched
chain (EC:1.17.99.3); K10214 3alpha,7alpha,12alpha-trihydroxy-5beta-cholestanoyl-CoA
24-hydroxylase [EC:1.17.99.3]
Length=681
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 33/88 (37%), Positives = 56/88 (63%), Gaps = 1/88 (1%)
Query 1 VTLLHIGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTRSRKLLYSTLTF 60
+T+ IG K+G++ +DNG L L +VR+PR+N+L RF EV DGTY+ + + Y +
Sbjct 236 ITVGDIGPKMGFENIDNGFLRLNHVRVPRENMLSRFAEVLPDGTYQRLGTPQSNYLGMLV 295
Query 61 TRKQII-RSAGAHLSRSIVVAIRYSAVR 87
TR Q++ + L ++ +A+RY+ +R
Sbjct 296 TRVQLLYKGFLPTLQKACTIAVRYAVIR 323
> ath:AT5G65110 ACX2; ACX2 (ACYL-COA OXIDASE 2); acyl-CoA oxidase;
K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=646
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 38/93 (40%), Positives = 53/93 (56%), Gaps = 5/93 (5%)
Query 1 VTLLHIGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTRS--RKLLYST- 57
V + G K+G GVDNG+L ++VRIPR NLL RF +V RDGTY ++ K +T
Sbjct 274 VEIQDCGHKVGLNGVDNGALRFRSVRIPRDNLLNRFGDVSRDGTYTSSLPTINKRFGATL 333
Query 58 --LTFTRKQIIRSAGAHLSRSIVVAIRYSAVRR 88
L R + ++ L S +AIRYS +R+
Sbjct 334 GELVGGRVGLAYASVGVLKISATIAIRYSLLRQ 366
> dre:569104 si:ch211-205h19.1
Length=652
Score = 58.9 bits (141), Expect = 4e-09, Method: Composition-based stats.
Identities = 36/92 (39%), Positives = 49/92 (53%), Gaps = 4/92 (4%)
Query 1 VTLLHIGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTRSR----KLLYS 56
VT+ + K G GVDNG L NVRIPR+NLL +F V DG Y + R + +
Sbjct 224 VTVSDMKHKEGLHGVDNGILIFDNVRIPRENLLGKFGSVSADGLYSSPALRGNRFNAMLA 283
Query 57 TLTFTRKQIIRSAGAHLSRSIVVAIRYSAVRR 88
LT TR + A A + +++A+ YS RR
Sbjct 284 ALTPTRLGLTVQAMAAMKLGLIIAVCYSHSRR 315
> hsa:8309 ACOX2, BCOX, BRCACOX, BRCOX, THCCox; acyl-CoA oxidase
2, branched chain (EC:1.17.99.3); K10214 3alpha,7alpha,12alpha-trihydroxy-5beta-cholestanoyl-CoA
24-hydroxylase [EC:1.17.99.3]
Length=681
Score = 58.5 bits (140), Expect = 4e-09, Method: Composition-based stats.
Identities = 31/84 (36%), Positives = 49/84 (58%), Gaps = 1/84 (1%)
Query 6 IGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTRSRKLLYSTLTFTRKQI 65
IG K+ + DNG L L +VR+PR+N+L RF +V DGTY + + Y + R ++
Sbjct 241 IGPKMDFDQTDNGFLQLNHVRVPRENMLSRFAQVLPDGTYVKLGTAQSNYLPMVVVRVEL 300
Query 66 IRSAGAH-LSRSIVVAIRYSAVRR 88
+ L ++ V+A+RYS +RR
Sbjct 301 LSGEILPILQKACVIAMRYSVIRR 324
> mmu:74121 Acoxl, 1200014P05Rik, AV025673; acyl-Coenzyme A oxidase-like
Length=632
Score = 58.2 bits (139), Expect = 6e-09, Method: Composition-based stats.
Identities = 36/93 (38%), Positives = 49/93 (52%), Gaps = 5/93 (5%)
Query 1 VTLLHIGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTRSRK-----LLY 55
VT + + K G GVDNG L VRIPR+NLL +F V DG Y + K +
Sbjct 199 VTAIDMMHKEGMNGVDNGILIFDKVRIPRENLLDKFGSVTPDGQYHSPIQSKNARFNAIL 258
Query 56 STLTFTRKQIIRSAGAHLSRSIVVAIRYSAVRR 88
+TLT +R + A + +++AIRYS RR
Sbjct 259 ATLTPSRLAVTFQALGAMKLGLMIAIRYSHSRR 291
> dre:406421 acox3, zgc:64087; acyl-Coenzyme A oxidase 3, pristanoyl
(EC:1.3.3.6); K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=692
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 33/88 (37%), Positives = 48/88 (54%), Gaps = 5/88 (5%)
Query 6 IGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRT-----TRSRKLLYSTLTF 60
+G+K+G G+DNG NVRIPR+NLL + +V DG Y + + L+
Sbjct 239 MGKKLGQNGLDNGFAVFHNVRIPRENLLNKTGDVAPDGQYVSPFKDPNKRFGASLGALSG 298
Query 61 TRKQIIRSAGAHLSRSIVVAIRYSAVRR 88
R I R A +L ++ VA+R+SA RR
Sbjct 299 GRVGITRMALVNLKLAVTVAVRFSATRR 326
> mmu:80911 Acox3, BI685180, EST-s59, PCOX; acyl-Coenzyme A oxidase
3, pristanoyl (EC:1.3.3.6); K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=700
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 33/87 (37%), Positives = 48/87 (55%), Gaps = 5/87 (5%)
Query 6 IGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTY----RTTRSR-KLLYSTLTF 60
IG+K+G G+DNG VRIPR+NLL R + +GTY + R R +L+
Sbjct 247 IGKKLGQNGLDNGFAMFNKVRIPRQNLLDRTGNITSEGTYNSPFKDVRQRLGASLGSLSS 306
Query 61 TRKQIIRSAGAHLSRSIVVAIRYSAVR 87
R II + +L ++ +AIR+SA R
Sbjct 307 GRISIISMSVVNLKLAVSIAIRFSATR 333
> ath:AT1G06310 ACX6; ACX6 (ACYL-COA OXIDASE 6); FAD binding /
acyl-CoA dehydrogenase/ acyl-CoA oxidase/ electron carrier/
oxidoreductase/ oxidoreductase, acting on the CH-CH group of
donors (EC:1.3.3.6); K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=675
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 36/93 (38%), Positives = 46/93 (49%), Gaps = 5/93 (5%)
Query 1 VTLLHIGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTT-----RSRKLLY 55
V + G KIG GVDNG + N+RIPR+NLL +V DG Y ++ +
Sbjct 270 VRIADCGHKIGLNGVDNGRIWFDNLRIPRENLLNSVADVLADGKYVSSIKDPDQRFGAFL 329
Query 56 STLTFTRKQIIRSAGAHLSRSIVVAIRYSAVRR 88
+ LT R I SA + VAIRYS RR
Sbjct 330 APLTSGRVTIASSAIYSAKLGLAVAIRYSLSRR 362
> ath:AT1G06290 ACX3; ACX3 (ACYL-COA OXIDASE 3); acyl-CoA oxidase
(EC:1.3.3.6); K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=675
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 34/87 (39%), Positives = 44/87 (50%), Gaps = 5/87 (5%)
Query 7 GQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTT-----RSRKLLYSTLTFT 61
G KIG GVDNG + N+RIPR+NLL +V DG Y ++ + + LT
Sbjct 276 GHKIGLNGVDNGRIWFDNLRIPRENLLNAVADVSSDGKYVSSIKDPDQRFGAFMAPLTSG 335
Query 62 RKQIIRSAGAHLSRSIVVAIRYSAVRR 88
R I SA + +AIRYS RR
Sbjct 336 RVTIASSAIYSAKVGLSIAIRYSLSRR 362
> hsa:8310 ACOX3; acyl-CoA oxidase 3, pristanoyl (EC:1.3.3.6);
K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=624
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 32/88 (36%), Positives = 50/88 (56%), Gaps = 5/88 (5%)
Query 6 IGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTY----RTTRSR-KLLYSTLTF 60
IG+K+G G+DNG VR+PR++LL R +V +GTY + R R +L+
Sbjct 247 IGKKLGQNGLDNGFAMFHKVRVPRQSLLNRMGDVTPEGTYVSPFKDVRQRFGASLGSLSS 306
Query 61 TRKQIIRSAGAHLSRSIVVAIRYSAVRR 88
R I+ A +L ++ +A+R+SA RR
Sbjct 307 GRVSIVSLAILNLKLAVAIALRFSATRR 334
> hsa:55289 ACOXL, FLJ11042; acyl-CoA oxidase-like
Length=580
Score = 54.3 bits (129), Expect = 8e-08, Method: Composition-based stats.
Identities = 35/93 (37%), Positives = 47/93 (50%), Gaps = 5/93 (5%)
Query 1 VTLLHIGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTTRSRK-----LLY 55
VT + + K G GVDNG L VRIPR+NLL +F V DG Y + K +
Sbjct 173 VTAIDMMYKEGLHGVDNGILIFDKVRIPRENLLDKFGSVAPDGQYHSPIRNKSARFNAML 232
Query 56 STLTFTRKQIIRSAGAHLSRSIVVAIRYSAVRR 88
+ LT +R + A + + +AIRYS RR
Sbjct 233 AALTPSRLAVAFQAMGAMKLGLTIAIRYSHSRR 265
> cel:F58F9.7 hypothetical protein; K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=667
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 32/94 (34%), Positives = 48/94 (51%), Gaps = 6/94 (6%)
Query 1 VTLLHIGQKIG-YQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRT-----TRSRKLL 54
+T+ +G K G +QGV+NG + KN R P LL + C++ DG Y T + + +
Sbjct 221 ITIGDMGSKPGCWQGVENGWMEFKNHRAPLSALLNKGCDITPDGKYVTSFKSASEKQSVS 280
Query 55 YSTLTFTRKQIIRSAGAHLSRSIVVAIRYSAVRR 88
TL+ R II + + +AIRYS RR
Sbjct 281 LGTLSVGRLGIIAKGMMACTFASTIAIRYSVARR 314
> sce:YGL205W POX1, FOX1; Fatty-acyl coenzyme A oxidase, involved
in the fatty acid beta-oxidation pathway; localized to the
peroxisomal matrix (EC:1.3.3.6); K00232 acyl-CoA oxidase
[EC:1.3.3.6]
Length=748
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 31/94 (32%), Positives = 50/94 (53%), Gaps = 6/94 (6%)
Query 1 VTLLHIGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDR--DGTYRTTRSRKLL---- 54
V++ IG K+G G+DNG + +NV IPR+ +L RF +V R DG+ +L
Sbjct 271 VSIGDIGAKMGRDGIDNGWIQFRNVVIPREFMLSRFTKVVRSPDGSVTVKTEPQLDQISG 330
Query 55 YSTLTFTRKQIIRSAGAHLSRSIVVAIRYSAVRR 88
YS L R ++ + S+ +A+RY+ R+
Sbjct 331 YSALLSGRVNMVMDSFRFGSKFATIAVRYAVGRQ 364
> tgo:TGME49_115480 acyl-CoA oxidase, putative (EC:1.9.3.1)
Length=856
Score = 42.4 bits (98), Expect = 3e-04, Method: Composition-based stats.
Identities = 20/43 (46%), Positives = 27/43 (62%), Gaps = 0/43 (0%)
Query 1 VTLLHIGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDG 43
VT+ +G +IG GVDN + NVRIPR +LL +V+ DG
Sbjct 146 VTIEDMGHRIGCNGVDNAKITFHNVRIPRSHLLDGITKVEADG 188
> cpv:cgd8_1270 clathrin heavy chain ; K04646 clathrin heavy chain
Length=2007
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 21/69 (30%), Positives = 32/69 (46%), Gaps = 12/69 (17%)
Query 3 LLHIGQKIGYQGVDNGSLHLKNVRIPRKNL--LMRFCEVDRDGTYRT----------TRS 50
LL Q+ GYQG D S+ + N I R++ R ++++G+ RT S
Sbjct 385 LLRWTQRYGYQGTDEFSIRMFNESIKRQDYQNACRVVSLNKNGSLRTPATLNHFKMVDSS 444
Query 51 RKLLYSTLT 59
KLL+ T
Sbjct 445 NKLLFQYFT 453
> mmu:71389 Chd6, 5430439G14Rik, 6330406J24Rik; chromodomain helicase
DNA binding protein 6; K14436 chromodomain-helicase-DNA-binding
protein 6 [EC:3.6.4.12]
Length=2711
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 18/35 (51%), Gaps = 0/35 (0%)
Query 33 LMRFCEVDRDGTYRTTRSRKLLYSTLTFTRKQIIR 67
LM F E+D D R TRSR+L + R + R
Sbjct 1064 LMEFSELDSDSDERPTRSRRLSDRARRYLRAECFR 1098
> hsa:84181 CHD6, CHD5, KIAA1335, RIGB; chromodomain helicase
DNA binding protein 6 (EC:3.6.4.12); K14436 chromodomain-helicase-DNA-binding
protein 6 [EC:3.6.4.12]
Length=2715
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 18/35 (51%), Gaps = 0/35 (0%)
Query 33 LMRFCEVDRDGTYRTTRSRKLLYSTLTFTRKQIIR 67
LM F E+D D R TRSR+L + R + R
Sbjct 1065 LMEFSELDSDSDERPTRSRRLNDKARRYLRAECFR 1099
> dre:100332228 nucleotide-binding oligomerization domain containing
2-like
Length=953
Score = 28.9 bits (63), Expect = 3.7, Method: Composition-based stats.
Identities = 17/38 (44%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 6 IGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDG 43
IG K+G GV S LK+ KNL+MR C V +G
Sbjct 749 IGNKLGDSGVKLLSDGLKDPHCKLKNLMMRDCGVTDEG 786
> hsa:51808 PHAX, FLJ13193, RNUXA; phosphorylated adaptor for
RNA export; K14291 phosphorylated adapter RNA export protein
Length=394
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 17/45 (37%), Positives = 25/45 (55%), Gaps = 1/45 (2%)
Query 4 LHIGQKIGYQGVDNGSLHLKNVRIPRKNLLMRFCEVDRDGTYRTT 48
+H G+K+G + +NG HLK R P K+ L E++ G Y T
Sbjct 179 MHGGKKMGSKEEENGQGHLKRKR-PVKDRLGNRPEMNYKGRYEIT 222
Lambda K H
0.327 0.139 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2021645584
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40