bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1497_orf2
Length=160
Score E
Sequences producing significant alignments: (Bits) Value
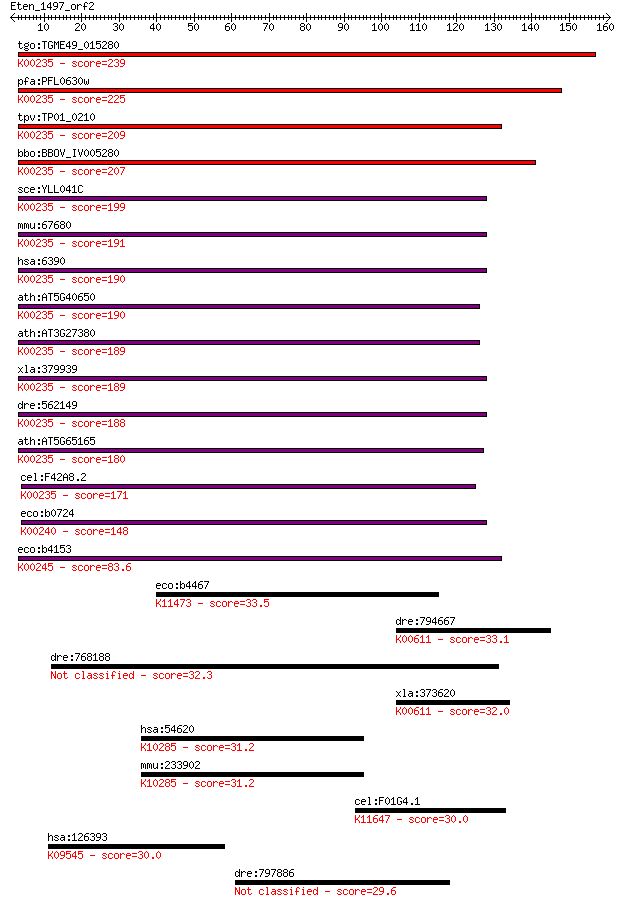
tgo:TGME49_015280 iron-sulfur subunit of succinate dehydrogena... 239 2e-63
pfa:PFL0630w iron-sulfur subunit of succinate dehydrogenase (E... 225 5e-59
tpv:TP01_0210 succinate dehydrogenase iron-sulfur subunit; K00... 209 2e-54
bbo:BBOV_IV005280 23.m06368; succinate dehydrogenase iron-sulf... 207 1e-53
sce:YLL041C SDH2, ACN17; Iron-sulfur protein subunit of succin... 199 2e-51
mmu:67680 Sdhb, 0710008N11Rik; succinate dehydrogenase complex... 191 1e-48
hsa:6390 SDHB, FLJ92337, IP, PGL4, SDH, SDH1, SDH2, SDHIP; suc... 190 1e-48
ath:AT5G40650 SDH2-2; SDH2-2; electron carrier/ succinate dehy... 190 2e-48
ath:AT3G27380 SDH2-1; SDH2-1; electron carrier/ succinate dehy... 189 3e-48
xla:379939 sdhb, Ip, MGC130803, MGC53699; succinate dehydrogen... 189 3e-48
dre:562149 sdhb, Ip, MGC165456, fj46d06, wu:fj46d06; succinate... 188 7e-48
ath:AT5G65165 SDH2-3; SDH2-3; electron carrier/ succinate dehy... 180 2e-45
cel:F42A8.2 sdhb-1; Succinate DeHydrogenase complex subunit B ... 171 9e-43
eco:b0724 sdhB, ECK0713, JW0714; succinate dehydrogenase, FeS ... 148 8e-36
eco:b4153 frdB, ECK4149, JW4114; fumarate reductase (anaerobic... 83.6 3e-16
eco:b4467 glcF, ECK2972, gox, JW5486, yghL; glycolate oxidase ... 33.5 0.28
dre:794667 otc, si:dkey-19h21.3; ornithine carbamoyltransferas... 33.1 0.40
dre:768188 zgc:154125 32.3 0.70
xla:373620 otc, otc-A, xotc; ornithine carbamoyltransferase (E... 32.0 0.91
hsa:54620 FBXL19, DKFZp434K0410, Fbl19, JHDM1C, MGC50505; F-bo... 31.2 1.5
mmu:233902 Fbxl19, 6030430O11, BC059812, Fbl19; F-box and leuc... 31.2 1.7
cel:F01G4.1 psa-4; Phasmid Socket Absent family member (psa-4)... 30.0 3.5
hsa:126393 HSPB6, FLJ32389, Hsp20; heat shock protein, alpha-c... 30.0 3.7
dre:797886 hypothetical LOC797886 29.6 4.9
> tgo:TGME49_015280 iron-sulfur subunit of succinate dehydrogenase,
putative (EC:1.3.5.1); K00235 succinate dehydrogenase
(ubiquinone) iron-sulfur protein [EC:1.3.5.1]
Length=342
Score = 239 bits (611), Expect = 2e-63, Method: Compositional matrix adjust.
Identities = 111/155 (71%), Positives = 131/155 (84%), Gaps = 1/155 (0%)
Query 3 NFYTQYKSIQPWLQRKTEPKEGS-EILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQ 61
NFY QY+S++PWL+RKT K+ + E LQSIEDR KLDG+YECILCACCSTSCPSYWWNPQ
Sbjct 186 NFYAQYRSVEPWLKRKTPKKDPNVENLQSIEDRRKLDGMYECILCACCSTSCPSYWWNPQ 245
Query 62 SYLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIR 121
+YLGPAVLMQAFRWI+DSRDEFT ERLA INDTMKLYRCHGIMNC++SCPKGL+PAGAI+
Sbjct 246 AYLGPAVLMQAFRWIADSRDEFTEERLAAINDTMKLYRCHGIMNCTVSCPKGLNPAGAIQ 305
Query 122 KMKKRIEEEFGTDFVKLAAQAARSRFLELAKELKI 156
KMK ++E F + L AQ A + +LAK+L +
Sbjct 306 KMKDQVEARFSPAWKSLQAQQALEKSQKLAKDLGL 340
> pfa:PFL0630w iron-sulfur subunit of succinate dehydrogenase
(EC:1.3.5.1); K00235 succinate dehydrogenase (ubiquinone) iron-sulfur
protein [EC:1.3.5.1]
Length=321
Score = 225 bits (573), Expect = 5e-59, Method: Compositional matrix adjust.
Identities = 99/146 (67%), Positives = 119/146 (81%), Gaps = 1/146 (0%)
Query 3 NFYTQYKSIQPWLQRKTEPKEG-SEILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQ 61
NFY QYKSI PWL+RKT+ ++G E QSIEDR KLDGLYECI+CA CSTSCPSYWWNP+
Sbjct 172 NFYNQYKSIDPWLKRKTKKEKGQKEFYQSIEDRKKLDGLYECIMCASCSTSCPSYWWNPE 231
Query 62 SYLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIR 121
YLGPA LMQA+RWI DSRDE+T+ERL ++NDTMKLYRCHGIMNC++ CPKGLDPA AI+
Sbjct 232 YYLGPATLMQAYRWIVDSRDEYTKERLMEVNDTMKLYRCHGIMNCTMCCPKGLDPAKAIK 291
Query 122 KMKKRIEEEFGTDFVKLAAQAARSRF 147
MK ++E F D +K +Q +S+
Sbjct 292 DMKNLVQENFSEDTIKEHSQYIKSKM 317
> tpv:TP01_0210 succinate dehydrogenase iron-sulfur subunit; K00235
succinate dehydrogenase (ubiquinone) iron-sulfur protein
[EC:1.3.5.1]
Length=263
Score = 209 bits (533), Expect = 2e-54, Method: Compositional matrix adjust.
Identities = 97/134 (72%), Positives = 112/134 (83%), Gaps = 6/134 (4%)
Query 3 NFYTQYKSIQPWLQRKTEPK----EGS-EILQSIEDRAKLDGLYECILCACCSTSCPSYW 57
NFY QY+S++PWL+RKT PK EG E QS EDR KLDGLYECILCACCSTSCPSYW
Sbjct 126 NFYEQYRSVEPWLKRKT-PKVLSEEGDKEYYQSREDRQKLDGLYECILCACCSTSCPSYW 184
Query 58 WNPQSYLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPA 117
WNP+ YLGPA LMQA+RWI+DSRDE+T ERL ++NDTMKLYRCHGI+NC+ CPKGLDPA
Sbjct 185 WNPEHYLGPAALMQAYRWIADSRDEYTTERLVEVNDTMKLYRCHGILNCTKVCPKGLDPA 244
Query 118 GAIRKMKKRIEEEF 131
G+I K+KK +EE
Sbjct 245 GSISKLKKLVEENV 258
> bbo:BBOV_IV005280 23.m06368; succinate dehydrogenase iron-sulfur
subunit (EC:1.3.5.1); K00235 succinate dehydrogenase (ubiquinone)
iron-sulfur protein [EC:1.3.5.1]
Length=281
Score = 207 bits (526), Expect = 1e-53, Method: Compositional matrix adjust.
Identities = 92/139 (66%), Positives = 114/139 (82%), Gaps = 2/139 (1%)
Query 3 NFYTQYKSIQPWLQRKTEPKEG-SEILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQ 61
NFY QY+++QPWL+RKT PK G +E LQS EDRA LDG+YECILCACC+TSCPSYWWNP+
Sbjct 127 NFYEQYRTVQPWLKRKT-PKSGEAEFLQSREDRALLDGMYECILCACCTTSCPSYWWNPE 185
Query 62 SYLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIR 121
Y+GPA LMQA+RWI DSRDE+T ER+ +ND+MKLYRCHGI+NC+ +CPKGLDPA AI
Sbjct 186 HYIGPAALMQAYRWIEDSRDEYTVERMVDVNDSMKLYRCHGILNCTRACPKGLDPAKAIS 245
Query 122 KMKKRIEEEFGTDFVKLAA 140
K+K ++E + +AA
Sbjct 246 KLKAKVEASADDTWRSIAA 264
> sce:YLL041C SDH2, ACN17; Iron-sulfur protein subunit of succinate
dehydrogenase (Sdh1p, Sdh2p, Sdh3p, Sdh4p), which couples
the oxidation of succinate to the transfer of electrons
to ubiquinone as part of the TCA cycle and the mitochondrial
respiratory chain (EC:1.3.5.1); K00235 succinate dehydrogenase
(ubiquinone) iron-sulfur protein [EC:1.3.5.1]
Length=266
Score = 199 bits (507), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 88/125 (70%), Positives = 107/125 (85%), Gaps = 0/125 (0%)
Query 3 NFYTQYKSIQPWLQRKTEPKEGSEILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQS 62
NFY QYKSIQP+LQR + PK+G+E+LQSIEDR KLDGLYECILCACCSTSCPSYWWN +
Sbjct 139 NFYQQYKSIQPYLQRSSFPKDGTEVLQSIEDRKKLDGLYECILCACCSTSCPSYWWNQEQ 198
Query 63 YLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIRK 122
YLGPAVLMQA+RW+ DSRD+ T+ R A +N++M LYRCH IMNC+ +CPKGL+P AI +
Sbjct 199 YLGPAVLMQAYRWLIDSRDQATKTRKAMLNNSMSLYRCHTIMNCTRTCPKGLNPGLAIAE 258
Query 123 MKKRI 127
+KK +
Sbjct 259 IKKSL 263
> mmu:67680 Sdhb, 0710008N11Rik; succinate dehydrogenase complex,
subunit B, iron sulfur (Ip) (EC:1.3.5.1); K00235 succinate
dehydrogenase (ubiquinone) iron-sulfur protein [EC:1.3.5.1]
Length=282
Score = 191 bits (484), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 86/126 (68%), Positives = 104/126 (82%), Gaps = 1/126 (0%)
Query 3 NFYTQYKSIQPWLQRKTEPKEGSE-ILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQ 61
NFY QYKSI+P+L++K E +EG + LQSIEDR KLDGLYECILCACCSTSCPSYWWN
Sbjct 147 NFYAQYKSIEPYLKKKDESQEGKQQYLQSIEDREKLDGLYECILCACCSTSCPSYWWNGD 206
Query 62 SYLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIR 121
YLGPAVLMQA+RW+ DSRD+FT ERLA++ D +YRCH IMNC+ +CPKGL+P AI
Sbjct 207 KYLGPAVLMQAYRWMIDSRDDFTEERLAKLQDPFSVYRCHTIMNCTQTCPKGLNPGKAIA 266
Query 122 KMKKRI 127
++KK +
Sbjct 267 EIKKMM 272
> hsa:6390 SDHB, FLJ92337, IP, PGL4, SDH, SDH1, SDH2, SDHIP; succinate
dehydrogenase complex, subunit B, iron sulfur (Ip)
(EC:1.3.5.1); K00235 succinate dehydrogenase (ubiquinone) iron-sulfur
protein [EC:1.3.5.1]
Length=280
Score = 190 bits (483), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 86/126 (68%), Positives = 104/126 (82%), Gaps = 1/126 (0%)
Query 3 NFYTQYKSIQPWLQRKTEPKEGSE-ILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQ 61
NFY QYKSI+P+L++K E +EG + LQSIE+R KLDGLYECILCACCSTSCPSYWWN
Sbjct 145 NFYAQYKSIEPYLKKKDESQEGKQQYLQSIEEREKLDGLYECILCACCSTSCPSYWWNGD 204
Query 62 SYLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIR 121
YLGPAVLMQA+RW+ DSRD+FT ERLA++ D LYRCH IMNC+ +CPKGL+P AI
Sbjct 205 KYLGPAVLMQAYRWMIDSRDDFTEERLAKLQDPFSLYRCHTIMNCTRTCPKGLNPGKAIA 264
Query 122 KMKKRI 127
++KK +
Sbjct 265 EIKKMM 270
> ath:AT5G40650 SDH2-2; SDH2-2; electron carrier/ succinate dehydrogenase
(EC:1.3.5.1); K00235 succinate dehydrogenase (ubiquinone)
iron-sulfur protein [EC:1.3.5.1]
Length=280
Score = 190 bits (482), Expect = 2e-48, Method: Compositional matrix adjust.
Identities = 86/124 (69%), Positives = 101/124 (81%), Gaps = 1/124 (0%)
Query 3 NFYTQYKSIQPWLQRKTEPK-EGSEILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQ 61
NFY QYKSI+PWL+RK G EILQS +DRAKLDG+YECILCACCSTSCPSYWWNP+
Sbjct 152 NFYNQYKSIEPWLKRKNPASVPGKEILQSKKDRAKLDGMYECILCACCSTSCPSYWWNPE 211
Query 62 SYLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIR 121
SYLGPA L+ A RWISDSRDE+T+ERL I+D KLYRCH I+NC+ +CPKGL+P I
Sbjct 212 SYLGPAALLHANRWISDSRDEYTKERLEAIDDEFKLYRCHTILNCARACPKGLNPGKQIT 271
Query 122 KMKK 125
+K+
Sbjct 272 HIKQ 275
> ath:AT3G27380 SDH2-1; SDH2-1; electron carrier/ succinate dehydrogenase
(EC:1.3.5.1); K00235 succinate dehydrogenase (ubiquinone)
iron-sulfur protein [EC:1.3.5.1]
Length=279
Score = 189 bits (481), Expect = 3e-48, Method: Compositional matrix adjust.
Identities = 86/124 (69%), Positives = 101/124 (81%), Gaps = 1/124 (0%)
Query 3 NFYTQYKSIQPWLQRKTEPK-EGSEILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQ 61
NFY QYKSI+PWL+RKT EILQS +DRAKLDG+YECILCACCSTSCPSYWWNP+
Sbjct 153 NFYNQYKSIEPWLKRKTPASVPAKEILQSKKDRAKLDGMYECILCACCSTSCPSYWWNPE 212
Query 62 SYLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIR 121
SYLGPA L+ A RWISDSRDE+T+ERL I+D KLYRCH I+NC+ +CPKGL+P I
Sbjct 213 SYLGPAALLHANRWISDSRDEYTKERLEAIDDEFKLYRCHTILNCARACPKGLNPGKQIT 272
Query 122 KMKK 125
+K+
Sbjct 273 HIKQ 276
> xla:379939 sdhb, Ip, MGC130803, MGC53699; succinate dehydrogenase
complex, subunit B, iron sulfur (Ip) (EC:1.3.5.1); K00235
succinate dehydrogenase (ubiquinone) iron-sulfur protein
[EC:1.3.5.1]
Length=282
Score = 189 bits (480), Expect = 3e-48, Method: Compositional matrix adjust.
Identities = 85/126 (67%), Positives = 104/126 (82%), Gaps = 1/126 (0%)
Query 3 NFYTQYKSIQPWLQRKTEPKEGSE-ILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQ 61
NFY QYKSI+P+L++K + ++G E LQSIEDR KLDGLYECILCACCSTSCPSYWWN
Sbjct 147 NFYAQYKSIEPYLKKKDKSQQGKEQYLQSIEDRDKLDGLYECILCACCSTSCPSYWWNAD 206
Query 62 SYLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIR 121
YLGPAVLMQA+RW+ DSRD+FT ERL+++ D LYRCH IMNC+ +CPKGL+P AI
Sbjct 207 KYLGPAVLMQAYRWMIDSRDDFTEERLSKLQDPFSLYRCHTIMNCTRTCPKGLNPGKAIA 266
Query 122 KMKKRI 127
++KK +
Sbjct 267 EIKKMM 272
> dre:562149 sdhb, Ip, MGC165456, fj46d06, wu:fj46d06; succinate
dehydrogenase complex, subunit B, iron sulfur (Ip) (EC:1.3.5.1);
K00235 succinate dehydrogenase (ubiquinone) iron-sulfur
protein [EC:1.3.5.1]
Length=280
Score = 188 bits (477), Expect = 7e-48, Method: Compositional matrix adjust.
Identities = 83/126 (65%), Positives = 104/126 (82%), Gaps = 1/126 (0%)
Query 3 NFYTQYKSIQPWLQRKTEPKEGSE-ILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQ 61
NFY QYKSI+P+L++K E ++G + LQS+EDR KLDGLYECILCACCSTSCPSYWWN
Sbjct 144 NFYAQYKSIEPYLKKKDESQQGKQQYLQSVEDRQKLDGLYECILCACCSTSCPSYWWNAD 203
Query 62 SYLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIR 121
YLGPAVLMQA+RW+ DSRD+FT +RL+++ D LYRCH IMNC+ +CPKGL+P AI
Sbjct 204 KYLGPAVLMQAYRWMIDSRDDFTEDRLSKLQDPFSLYRCHTIMNCTRTCPKGLNPGKAIA 263
Query 122 KMKKRI 127
++KK +
Sbjct 264 EIKKMM 269
> ath:AT5G65165 SDH2-3; SDH2-3; electron carrier/ succinate dehydrogenase
(EC:1.3.5.1); K00235 succinate dehydrogenase (ubiquinone)
iron-sulfur protein [EC:1.3.5.1]
Length=309
Score = 180 bits (456), Expect = 2e-45, Method: Compositional matrix adjust.
Identities = 80/125 (64%), Positives = 99/125 (79%), Gaps = 1/125 (0%)
Query 3 NFYTQYKSIQPWLQRKTEPKEGSEILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQS 62
NFY QYKS++PWL+ + PK+G E QS +DR KLDGLYECILCACC+TSCPSYWWNP+
Sbjct 172 NFYQQYKSMEPWLKTRKPPKDGREHRQSPKDRKKLDGLYECILCACCTTSCPSYWWNPEE 231
Query 63 YLGPAVLMQAFRWISDSRDEFTRERLAQINDT-MKLYRCHGIMNCSISCPKGLDPAGAIR 121
+ GPA L+QA+RWISDSRDE+ ERL I ++ K+YRC I NC+ +CPKGL+PA AI
Sbjct 232 FPGPAALLQAYRWISDSRDEYREERLQAITESETKVYRCRAIKNCTATCPKGLNPASAIL 291
Query 122 KMKKR 126
KMK +
Sbjct 292 KMKSK 296
> cel:F42A8.2 sdhb-1; Succinate DeHydrogenase complex subunit
B family member (sdhb-1); K00235 succinate dehydrogenase (ubiquinone)
iron-sulfur protein [EC:1.3.5.1]
Length=298
Score = 171 bits (433), Expect = 9e-43, Method: Compositional matrix adjust.
Identities = 74/122 (60%), Positives = 96/122 (78%), Gaps = 1/122 (0%)
Query 4 FYTQYKSIQPWLQRKTEPKEGS-EILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQS 62
FY QY SIQPW+Q+KT G ++ QS+ +R +LDGLYECILCACCSTSCPSYWWN
Sbjct 160 FYAQYASIQPWIQKKTPLTLGEKQMHQSVAERDRLDGLYECILCACCSTSCPSYWWNADK 219
Query 63 YLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIRK 122
YLGPAVLMQA+RW+ DSRD++ ERL +++D+ ++CH IMNC+ +CPK L+PA AI +
Sbjct 220 YLGPAVLMQAYRWVIDSRDDYATERLHRMHDSFSAFKCHTIMNCTKTCPKHLNPAKAIGE 279
Query 123 MK 124
+K
Sbjct 280 IK 281
> eco:b0724 sdhB, ECK0713, JW0714; succinate dehydrogenase, FeS
subunit (EC:1.3.99.1); K00240 succinate dehydrogenase iron-sulfur
protein [EC:1.3.99.1]
Length=238
Score = 148 bits (373), Expect = 8e-36, Method: Compositional matrix adjust.
Identities = 66/124 (53%), Positives = 84/124 (67%), Gaps = 0/124 (0%)
Query 4 FYTQYKSIQPWLQRKTEPKEGSEILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQSY 63
FY QY+ I+P+L + E LQ E R KLDGLYECILCACCSTSCPS+WWNP +
Sbjct 110 FYAQYEKIKPYLLNNGQNPPAREHLQMPEQREKLDGLYECILCACCSTSCPSFWWNPDKF 169
Query 64 LGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIRKM 123
+GPA L+ A+R++ DSRD T RL ++D ++RCH IMNC CPKGL+P AI +
Sbjct 170 IGPAGLLAAYRFLIDSRDTETDSRLDGLSDAFSVFRCHSIMNCVSVCPKGLNPTRAIGHI 229
Query 124 KKRI 127
K +
Sbjct 230 KSML 233
> eco:b4153 frdB, ECK4149, JW4114; fumarate reductase (anaerobic),
Fe-S subunit (EC:1.3.99.1); K00245 fumarate reductase iron-sulfur
protein [EC:1.3.99.1]
Length=244
Score = 83.6 bits (205), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 41/129 (31%), Positives = 69/129 (53%), Gaps = 1/129 (0%)
Query 3 NFYTQYKSIQPWLQRKTEPKEGSEILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQS 62
+F ++I+P++ + + +Q+ AK CI C C +CP + NP+
Sbjct 109 HFIESLEAIKPYIIGNSRTADQGTNIQTPAQMAKYHQFSGCINCGLCYAACPQFGLNPE- 167
Query 63 YLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIRK 122
++GPA + A R+ DSRD +ER+AQ+N ++ C + CS CPK +DPA AI++
Sbjct 168 FIGPAAITLAHRYNEDSRDHGKKERMAQLNSQNGVWSCTFVGYCSEVCPKHVDPAAAIQQ 227
Query 123 MKKRIEEEF 131
K ++F
Sbjct 228 GKVESSKDF 236
> eco:b4467 glcF, ECK2972, gox, JW5486, yghL; glycolate oxidase
iron-sulfur subunit; K11473 glycolate oxidase iron-sulfur
subunit
Length=407
Score = 33.5 bits (75), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 19/75 (25%), Positives = 31/75 (41%), Gaps = 7/75 (9%)
Query 40 LYECILCACCSTSCPSYWWNPQSYLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYR 99
L C+ C C+ +CP+Y GP + + + + +E T + L R
Sbjct 22 LRACVHCGFCTATCPTYQLLGDELDGPRGRIYLIKQVLEG-NEVT------LKTQEHLDR 74
Query 100 CHGIMNCSISCPKGL 114
C NC +CP G+
Sbjct 75 CLTCRNCETTCPSGV 89
> dre:794667 otc, si:dkey-19h21.3; ornithine carbamoyltransferase;
K00611 ornithine carbamoyltransferase [EC:2.1.3.3]
Length=352
Score = 33.1 bits (74), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 26/43 (60%), Gaps = 2/43 (4%)
Query 104 MNCSISCPKGLDPAGAIRKMKKRIEEEFGT--DFVKLAAQAAR 144
+N ++ PKG +P A+ + +R+ ++FGT FV +AAR
Sbjct 211 VNLKVATPKGYEPDAAVTEEAQRLAKQFGTRLQFVSDPVEAAR 253
> dre:768188 zgc:154125
Length=425
Score = 32.3 bits (72), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 28/126 (22%), Positives = 48/126 (38%), Gaps = 24/126 (19%)
Query 12 QPWLQRKTEPKEGSEILQSIE-----DRAKLDGLYECILCAC--CSTSCPSYWWNPQSYL 64
++ KEG Q I+ AK+ + C+C CS C SY+W
Sbjct 263 HSYISATAAEKEGDSDAQEIKVIEGPPAAKMSASGHTLECSCPNCSDVCKSYYW------ 316
Query 65 GPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIRKMK 124
+ + D ++R RL++ + R +G+ C ++C KG + +
Sbjct 317 ----------YYTPFNDRYSRSRLSEYGKNI-YARENGLYCCRMNCGKGFSRFSQLYSHR 365
Query 125 KRIEEE 130
R E E
Sbjct 366 ARSESE 371
> xla:373620 otc, otc-A, xotc; ornithine carbamoyltransferase
(EC:2.1.3.3); K00611 ornithine carbamoyltransferase [EC:2.1.3.3]
Length=351
Score = 32.0 bits (71), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 18/30 (60%), Gaps = 0/30 (0%)
Query 104 MNCSISCPKGLDPAGAIRKMKKRIEEEFGT 133
MN I+ PKG +P A+ K+ +EFGT
Sbjct 210 MNLRIATPKGYEPDSAVISAAKQFSKEFGT 239
> hsa:54620 FBXL19, DKFZp434K0410, Fbl19, JHDM1C, MGC50505; F-box
and leucine-rich repeat protein 19; K10285 F-box and leucine-rich
repeat protein 19
Length=694
Score = 31.2 bits (69), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 27/63 (42%), Gaps = 12/63 (19%)
Query 36 KLDGLYECILCACCSTSCPSYWWNPQSYLG----PAVLMQAFRWISDSRDEFTRERLAQI 91
+L GL E +L C W S LG PA+ + RWI D +D RE L
Sbjct 506 RLQGLQELVLSGCS--------WLSVSALGSAPLPALRLLDLRWIEDVKDSQLRELLLPP 557
Query 92 NDT 94
DT
Sbjct 558 PDT 560
> mmu:233902 Fbxl19, 6030430O11, BC059812, Fbl19; F-box and leucine-rich
repeat protein 19; K10285 F-box and leucine-rich
repeat protein 19
Length=674
Score = 31.2 bits (69), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 27/63 (42%), Gaps = 12/63 (19%)
Query 36 KLDGLYECILCACCSTSCPSYWWNPQSYLG----PAVLMQAFRWISDSRDEFTRERLAQI 91
+L GL E +L C W S LG PA+ + RWI D +D RE L
Sbjct 486 RLQGLQELVLSGCS--------WLSVSALGSAPLPALRLLDLRWIEDVKDSQLRELLLPP 537
Query 92 NDT 94
DT
Sbjct 538 PDT 540
> cel:F01G4.1 psa-4; Phasmid Socket Absent family member (psa-4);
K11647 SWI/SNF-related matrix-associated actin-dependent
regulator of chromatin subfamily A member 2/4 [EC:3.6.4.-]
Length=1474
Score = 30.0 bits (66), Expect = 3.5, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 23/43 (53%), Gaps = 3/43 (6%)
Query 93 DTMKLYRCHGIM---NCSISCPKGLDPAGAIRKMKKRIEEEFG 132
D MK++ H I +IS P G+DP G +++ + I+ G
Sbjct 149 DLMKIFNLHQIRCNRPTTISVPSGIDPVGMLKQRENMIQNRIG 191
> hsa:126393 HSPB6, FLJ32389, Hsp20; heat shock protein, alpha-crystallin-related,
B6; K09545 heat shock 27kDa protein 6
Length=160
Score = 30.0 bits (66), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 24/48 (50%), Gaps = 1/48 (2%)
Query 11 IQP-WLQRKTEPKEGSEILQSIEDRAKLDGLYECILCACCSTSCPSYW 57
+QP WL+R + P G + D+ +GL E L A C T+ Y+
Sbjct 7 VQPSWLRRASAPLPGLSAPGRLFDQRFGEGLLEAELAALCPTTLAPYY 54
> dre:797886 hypothetical LOC797886
Length=213
Score = 29.6 bits (65), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 24/57 (42%), Gaps = 2/57 (3%)
Query 61 QSYLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPA 117
Q Y G + + D D R L I D + LYRC G N ISC + + A
Sbjct 155 QKYQGMKAFVYTNIYTEDQTDPELRAELKTIADLVPLYRCKG--NSCISCGQDVIDA 209
Lambda K H
0.321 0.135 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3712313100
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40