bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1391_orf1
Length=275
Score E
Sequences producing significant alignments: (Bits) Value
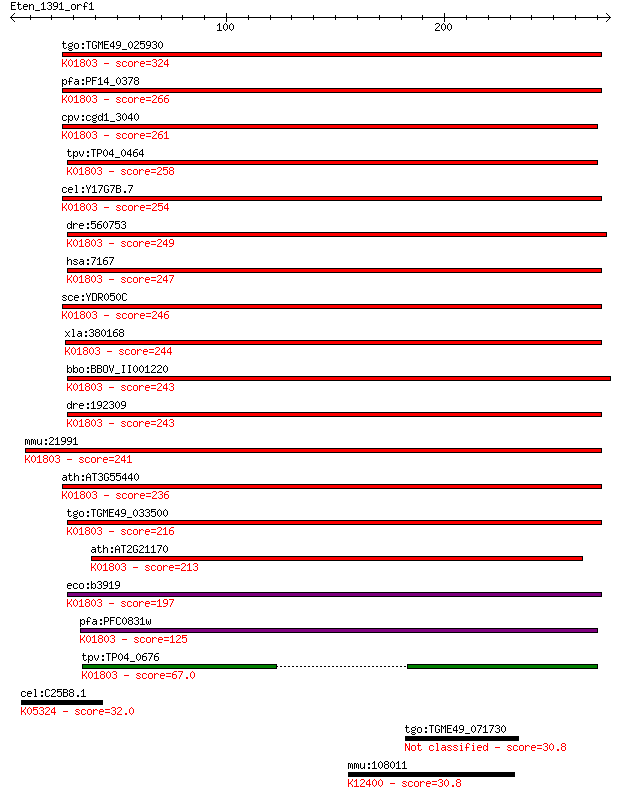
tgo:TGME49_025930 triosephosphate isomerase, putative (EC:5.3.... 324 2e-88
pfa:PF14_0378 triosephosphate isomerase (EC:5.3.1.1); K01803 t... 266 5e-71
cpv:cgd1_3040 triosephosphate isomerase (EC:5.3.1.1); K01803 t... 261 1e-69
tpv:TP04_0464 triosephosphate isomerase (EC:5.3.1.1); K01803 t... 258 2e-68
cel:Y17G7B.7 tpi-1; Triose Phosphate Isomerase family member (... 254 3e-67
dre:560753 tpi1b, DrTPI-B, MGC174772, MGC64167, cb179, wu:fb51... 249 7e-66
hsa:7167 TPI1, MGC88108, TPI; triosephosphate isomerase 1 (EC:... 247 4e-65
sce:YDR050C TPI1; Tpi1p (EC:5.3.1.1); K01803 triosephosphate i... 246 8e-65
xla:380168 tpi1, MGC52638, TIM, tpi; triosephosphate isomerase... 244 2e-64
bbo:BBOV_II001220 18.m06090; triosephosphate isomerase protein... 243 6e-64
dre:192309 tpi1a, DrTPI-A; triosephosphate isomerase 1a (EC:5.... 243 8e-64
mmu:21991 Tpi1, AI255506, Tpi, Tpi-1; triosephosphate isomeras... 241 2e-63
ath:AT3G55440 TPI; TPI (TRIOSEPHOSPHATE ISOMERASE); triose-pho... 236 9e-62
tgo:TGME49_033500 triosephosphate isomerase, putative (EC:5.3.... 216 8e-56
ath:AT2G21170 TIM; TIM (TRIOSEPHOSPHATE ISOMERASE); catalytic/... 213 4e-55
eco:b3919 tpiA, ECK3911, JW3890, tpi; triosephosphate isomeras... 197 3e-50
pfa:PFC0831w triosephophate isomerase, putative (EC:5.3.1.1); ... 125 1e-28
tpv:TP04_0676 triosephosphate isomerase (EC:5.3.1.1); K01803 t... 67.0 7e-11
cel:C25B8.1 kqt-1; potassium channel, KvQLT family member (kqt... 32.0 2.4
tgo:TGME49_071730 seryl-tRNA synthetase, putative (EC:6.1.1.11) 30.8 5.2
mmu:108011 Ap4e1, 2310033A20Rik, 9930028M04Rik, AV087807; adap... 30.8 5.6
> tgo:TGME49_025930 triosephosphate isomerase, putative (EC:5.3.1.1);
K01803 triosephosphate isomerase (TIM) [EC:5.3.1.1]
Length=253
Score = 324 bits (831), Expect = 2e-88, Method: Compositional matrix adjust.
Identities = 152/247 (61%), Positives = 188/247 (76%), Gaps = 0/247 (0%)
Query 25 MPRTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKNGFQ 84
M RTPW+GGNWK NGTV SIT+L E EFDP +VV+FP ALH L +EKL +
Sbjct 1 MVRTPWVGGNWKCNGTVGSITDLCGEFGKTEFDPKTIDVVIFPPALHAPLTREKLPKKYH 60
Query 85 IGCQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMKEE 144
+G QN SKT GAFTGEI+ M+KDFGL+W++ GHSERRQYY E+D VVAEKV + ++E+
Sbjct 61 VGLQNCSKTHNGAFTGEISVEMIKDFGLRWILAGHSERRQYYGESDEVVAEKVNIILQEK 120
Query 145 GIHVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVATPE 204
++V +C+GE+L++REANKT +V QL A LPK+SDW RVVIAYEPVWAIGTGKVATP
Sbjct 121 DLNVVLCVGEQLKDREANKTNDVVDAQLAACLPKISDWDRVVIAYEPVWAIGTGKVATPA 180
Query 205 QAQEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLKKA 264
QAQEVH +R+F+ K E VA ++RIVYGGSV+ N L+ D+DGFLVGGASLKK
Sbjct 181 QAQEVHEHIREFLKAKVSEDVANKVRIVYGGSVNASNSTELILQPDLDGFLVGGASLKKD 240
Query 265 FVDIIGA 271
F+DII +
Sbjct 241 FLDIIAS 247
> pfa:PF14_0378 triosephosphate isomerase (EC:5.3.1.1); K01803
triosephosphate isomerase (TIM) [EC:5.3.1.1]
Length=248
Score = 266 bits (681), Expect = 5e-71, Method: Compositional matrix adjust.
Identities = 118/247 (47%), Positives = 176/247 (71%), Gaps = 1/247 (0%)
Query 25 MPRTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKNGFQ 84
M R ++ NWK NGT+ SI L+ N+ +FDPS+ +VVVFP ++H ++ L++ F
Sbjct 1 MARKYFVAANWKCNGTLESIKSLTNSFNNLDFDPSKLDVVVFPVSVHYDHTRKLLQSKFS 60
Query 85 IGCQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMKEE 144
G QN+SK G G++TGE++A + KD +++V++GH ERR+Y+ ETD V EK+ ++K
Sbjct 61 TGIQNVSKFGNGSYTGEVSAEIAKDLNIEYVIIGHFERRKYFHETDEDVREKLQASLKN- 119
Query 145 GIHVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVATPE 204
+ +C GE LE+RE NKT+EV +Q++A + + ++ V++AYEP+WAIGTGK ATPE
Sbjct 120 NLKAVVCFGESLEQREQNKTIEVITKQVKAFVDLIDNFDNVILAYEPLWAIGTGKTATPE 179
Query 205 QAQEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLKKA 264
QAQ VH +RK V + GE A ++RI+YGGSV+ +NC +L++ D+DGFLVG ASLK++
Sbjct 180 QAQLVHKEIRKIVKDTCGEKQANQIRILYGGSVNTENCSSLIQQEDIDGFLVGNASLKES 239
Query 265 FVDIIGA 271
FVDII +
Sbjct 240 FVDIIKS 246
> cpv:cgd1_3040 triosephosphate isomerase (EC:5.3.1.1); K01803
triosephosphate isomerase (TIM) [EC:5.3.1.1]
Length=250
Score = 261 bits (668), Expect = 1e-69, Method: Compositional matrix adjust.
Identities = 132/249 (53%), Positives = 163/249 (65%), Gaps = 7/249 (2%)
Query 25 MPRTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKNG-- 82
M R ++GGN+K NGT S+ L E S +EV VFPT+LHI L KE N
Sbjct 1 MSRKYFVGGNFKCNGTKESLKTLIDSFKQVE--SSNSEVYVFPTSLHISLVKEFFGNDHP 58
Query 83 --FQIGCQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMA 140
F+IG QNIS TG GAFTGE++ M+KD + +VGHSERRQYYSETD +V KV
Sbjct 59 GVFKIGSQNISCTGNGAFTGEVSCEMLKDMDVDCSLVGHSERRQYYSETDQIVNNKVKKG 118
Query 141 MKEEGIHVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKV 200
+ E G+ + +CIGE L ERE KT +V ++QL L VSD +VIAYEP+WAIGTG V
Sbjct 119 L-ENGLKIVLCIGESLSERETGKTNDVIQKQLTEALKDVSDLSNLVIAYEPIWAIGTGVV 177
Query 201 ATPEQAQEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGAS 260
ATP QAQE HA +R++V V+ LRI+YGGSV+ NC L++C D+DGFLVGGAS
Sbjct 178 ATPGQAQEAHAFIREYVTRMYNPQVSSNLRIIYGGSVTPDNCNELIKCADIDGFLVGGAS 237
Query 261 LKKAFVDII 269
LK F II
Sbjct 238 LKPTFAKII 246
> tpv:TP04_0464 triosephosphate isomerase (EC:5.3.1.1); K01803
triosephosphate isomerase (TIM) [EC:5.3.1.1]
Length=250
Score = 258 bits (658), Expect = 2e-68, Method: Compositional matrix adjust.
Identities = 122/244 (50%), Positives = 172/244 (70%), Gaps = 3/244 (1%)
Query 27 RTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKNG-FQI 85
R W+GGNWK NG+ SI++L N+ + + +VV+FP +L++ + LK G F++
Sbjct 4 RRKWLGGNWKCNGSKQSISDLLAHFNNLKSKHNNLDVVLFPPSLYVEHTRNVLKTGVFEL 63
Query 86 GCQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMKEEG 145
G QN+S++ GAFTGE++ M DFGLKW +VGHSERRQ ++E + V KV M +++ G
Sbjct 64 GVQNVSQSKSGAFTGELSLPMFSDFGLKWSLVGHSERRQLFNEDNFDVRAKVKM-LQDNG 122
Query 146 IHVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVATPEQ 205
++ +C GE L ERE +T+ V ++QL A + V DW +VV+AYEPVWAIGTGKVAT EQ
Sbjct 123 VNAVVCFGETLSEREQGQTVNVLKRQLNAFVKDVKDWDKVVLAYEPVWAIGTGKVATVEQ 182
Query 206 AQEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLKKAF 265
+ H +R+ V AG AVA+++R+VYGGSV+E+NC L +C DVDGFLVGG SLKK F
Sbjct 183 VNQAHKFVREHVRGLAG-AVADKVRVVYGGSVNEKNCHELAKCNDVDGFLVGGTSLKKEF 241
Query 266 VDII 269
+D++
Sbjct 242 LDVL 245
> cel:Y17G7B.7 tpi-1; Triose Phosphate Isomerase family member
(tpi-1); K01803 triosephosphate isomerase (TIM) [EC:5.3.1.1]
Length=247
Score = 254 bits (649), Expect = 3e-67, Method: Compositional matrix adjust.
Identities = 133/247 (53%), Positives = 166/247 (67%), Gaps = 2/247 (0%)
Query 25 MPRTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKNGFQ 84
M R ++GGNWK NG AS+ + LN A D S +VVV P A ++ AK KLK G
Sbjct 1 MTRKFFVGGNWKMNGDYASVDGIVTFLN-ASADNSSVDVVVAPPAPYLAYAKSKLKAGVL 59
Query 85 IGCQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMKEE 144
+ QN K GAFTGEI+ M+KD GL+WV++GHSERR + E+D+++AEK A+ E
Sbjct 60 VAAQNCYKVPKGAFTGEISPAMIKDLGLEWVILGHSERRHVFGESDALIAEKTVHAL-EA 118
Query 145 GIHVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVATPE 204
GI V CIGEKLEEREA T +V +QL+A++ K W +VIAYEPVWAIGTGK A+ E
Sbjct 119 GIKVVFCIGEKLEEREAGHTKDVNFRQLQAIVDKGVSWENIVIAYEPVWAIGTGKTASGE 178
Query 205 QAQEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLKKA 264
QAQEVH +R F+ EK AVA+ RI+YGGSV+ N L + D+DGFLVGGASLK
Sbjct 179 QAQEVHEWIRAFLKEKVSPAVADATRIIYGGSVTADNAAELGKKPDIDGFLVGGASLKPD 238
Query 265 FVDIIGA 271
FV II A
Sbjct 239 FVKIINA 245
> dre:560753 tpi1b, DrTPI-B, MGC174772, MGC64167, cb179, wu:fb51c02;
triosephosphate isomerase 1b (EC:5.3.1.1); K01803 triosephosphate
isomerase (TIM) [EC:5.3.1.1]
Length=248
Score = 249 bits (636), Expect = 7e-66, Method: Compositional matrix adjust.
Identities = 126/247 (51%), Positives = 165/247 (66%), Gaps = 2/247 (0%)
Query 27 RTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKNGFQIG 86
R ++GGNWK NG SI EL+ LN A+ +P TEVV +++ A+ KL +
Sbjct 4 RKFFVGGNWKMNGDKKSIEELANTLNSAKLNPD-TEVVCGAPTIYLDYARSKLNPNIDVA 62
Query 87 CQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMKEEGI 146
QN K GAFTGEI+ M+KD G+KWV++GHSERR + E+D ++ +KVA A+ E G+
Sbjct 63 AQNCYKVAKGAFTGEISPAMIKDCGVKWVILGHSERRHVFGESDELIGQKVAHAL-ENGL 121
Query 147 HVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVATPEQA 206
V CIGEKL+EREA T +V Q + + V DW +VV+AYEPVWAIGTGK A+P+QA
Sbjct 122 GVIACIGEKLDEREAGITEKVVFAQTKFIADNVKDWSKVVLAYEPVWAIGTGKTASPQQA 181
Query 207 QEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLKKAFV 266
QEVH LR+++ EAVA +RI+YGGSV+ C+ L D+DGFLVGGASLK F+
Sbjct 182 QEVHDKLRQWLKTNVSEAVANSVRIIYGGSVTGGTCKELASQKDLDGFLVGGASLKPEFI 241
Query 267 DIIGAPA 273
DII A A
Sbjct 242 DIINAKA 248
> hsa:7167 TPI1, MGC88108, TPI; triosephosphate isomerase 1 (EC:5.3.1.1);
K01803 triosephosphate isomerase (TIM) [EC:5.3.1.1]
Length=249
Score = 247 bits (630), Expect = 4e-65, Method: Compositional matrix adjust.
Identities = 128/245 (52%), Positives = 163/245 (66%), Gaps = 2/245 (0%)
Query 27 RTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKNGFQIG 86
R ++GGNWK NG S+ EL LN A+ P+ TEVV P +I A++KL +
Sbjct 5 RKFFVGGNWKMNGRKQSLGELIGTLNAAKV-PADTEVVCAPPTAYIDFARQKLDPKIAVA 63
Query 87 CQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMKEEGI 146
QN K GAFTGEI+ GM+KD G WV++GHSERR + E+D ++ +KVA A+ E G+
Sbjct 64 AQNCYKVTNGAFTGEISPGMIKDCGATWVVLGHSERRHVFGESDELIGQKVAHALAE-GL 122
Query 147 HVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVATPEQA 206
V CIGEKL+EREA T +V +Q + + V DW +VV+AYEPVWAIGTGK ATP+QA
Sbjct 123 GVIACIGEKLDEREAGITEKVVFEQTKVIADNVKDWSKVVLAYEPVWAIGTGKTATPQQA 182
Query 207 QEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLKKAFV 266
QEVH LR ++ +AVA+ RI+YGGSV+ C+ L DVDGFLVGGASLK FV
Sbjct 183 QEVHEKLRGWLKSNVSDAVAQSTRIIYGGSVTGATCKELASQPDVDGFLVGGASLKPEFV 242
Query 267 DIIGA 271
DII A
Sbjct 243 DIINA 247
> sce:YDR050C TPI1; Tpi1p (EC:5.3.1.1); K01803 triosephosphate
isomerase (TIM) [EC:5.3.1.1]
Length=248
Score = 246 bits (627), Expect = 8e-65, Method: Compositional matrix adjust.
Identities = 125/248 (50%), Positives = 164/248 (66%), Gaps = 3/248 (1%)
Query 25 MPRTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKN-GF 83
M RT ++GGN+K NG+ SI E+ + LN A P EVV+ P A ++ + +K
Sbjct 1 MARTFFVGGNFKLNGSKQSIKEIVERLNTASI-PENVEVVICPPATYLDYSVSLVKKPQV 59
Query 84 QIGCQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMKE 143
+G QN GAFTGE + +KD G KWV++GHSERR Y+ E D +A+K A+ +
Sbjct 60 TVGAQNAYLKASGAFTGENSVDQIKDVGAKWVILGHSERRSYFHEDDKFIADKTKFALGQ 119
Query 144 EGIHVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVATP 203
G+ V +CIGE LEE++A KT++V +QL AVL +V DW VV+AYEPVWAIGTG ATP
Sbjct 120 -GVGVILCIGETLEEKKAGKTLDVVERQLNAVLEEVKDWTNVVVAYEPVWAIGTGLAATP 178
Query 204 EQAQEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLKK 263
E AQ++HA++RKF+A K G+ A ELRI+YGGS + N DVDGFLVGGASLK
Sbjct 179 EDAQDIHASIRKFLASKLGDKAASELRILYGGSANGSNAVTFKDKADVDGFLVGGASLKP 238
Query 264 AFVDIIGA 271
FVDII +
Sbjct 239 EFVDIINS 246
> xla:380168 tpi1, MGC52638, TIM, tpi; triosephosphate isomerase
1 (EC:5.3.1.1); K01803 triosephosphate isomerase (TIM) [EC:5.3.1.1]
Length=248
Score = 244 bits (623), Expect = 2e-64, Method: Compositional matrix adjust.
Identities = 125/246 (50%), Positives = 163/246 (66%), Gaps = 2/246 (0%)
Query 26 PRTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKNGFQI 85
PR ++GGNWK NG S+ EL LN + + + TEVV A+++ A++KL +
Sbjct 3 PRKFFVGGNWKMNGDKKSLGELINTLNSGKMN-ADTEVVCGAPAIYLDFARQKLDAKIAL 61
Query 86 GCQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMKEEG 145
QN K GAFTGEI+ M+KD G WV++GHSERR + E D ++ +KVA A+ EG
Sbjct 62 SAQNCYKVAKGAFTGEISPAMIKDCGATWVILGHSERRHVFGECDELIGQKVAHAL-SEG 120
Query 146 IHVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVATPEQ 205
I V CIGEKL++REA T +V +Q +A+ V DW +VV+AYEPVWAIGTGK ATPEQ
Sbjct 121 IGVIACIGEKLDQREAGITEKVVFEQTKAIADNVKDWSKVVLAYEPVWAIGTGKTATPEQ 180
Query 206 AQEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLKKAF 265
AQEVH LR++V E VA+ +RI+YGGSV+ C L D+DGFLVGGASLK F
Sbjct 181 AQEVHKKLREWVKTNVSEGVAQSVRIIYGGSVTGGTCRELAGQPDIDGFLVGGASLKPEF 240
Query 266 VDIIGA 271
++II A
Sbjct 241 IEIINA 246
> bbo:BBOV_II001220 18.m06090; triosephosphate isomerase protein
(EC:5.3.1.1); K01803 triosephosphate isomerase (TIM) [EC:5.3.1.1]
Length=253
Score = 243 bits (620), Expect = 6e-64, Method: Compositional matrix adjust.
Identities = 122/250 (48%), Positives = 167/250 (66%), Gaps = 3/250 (1%)
Query 27 RTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKL-KNGFQI 85
R W+GGNWK NG+ ++ L+K + +FD + +VV+FP+ L+I A E K F+I
Sbjct 5 RKRWVGGNWKCNGSFELVSTLAKAFDAFKFDGNAMDVVLFPSNLYITKALECFDKQRFKI 64
Query 86 GCQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMKEEG 145
G QN S+ GAFTGEI M+ + GL+W ++GHSERR + ETD++VA+KV A ++
Sbjct 65 GVQNFSQAKCGAFTGEIAIPMLNELGLEWTLIGHSERRTLFGETDAIVAQKVNTA-QQGK 123
Query 146 IHVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVATPEQ 205
+ +CIGE L ERE+ + V QL+A + V+DW VVIAYEPVWAIGTGKVAT E+
Sbjct 124 LRAAVCIGENLTERESGRVEAVLTTQLDAFMGMVTDWDIVVIAYEPVWAIGTGKVATCEE 183
Query 206 AQEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLKKAF 265
+E H +R ++ K G +VAE +RIVYGGSV+EQNC+ LL ++DGFLVGG S+ F
Sbjct 184 VREAHQMIRDYMTSKLG-SVAETIRIVYGGSVNEQNCQELLHVPNMDGFLVGGKSITPGF 242
Query 266 VDIIGAPARV 275
DI+ A V
Sbjct 243 ADIMEAAYNV 252
> dre:192309 tpi1a, DrTPI-A; triosephosphate isomerase 1a (EC:5.3.1.1);
K01803 triosephosphate isomerase (TIM) [EC:5.3.1.1]
Length=248
Score = 243 bits (619), Expect = 8e-64, Method: Compositional matrix adjust.
Identities = 124/245 (50%), Positives = 163/245 (66%), Gaps = 2/245 (0%)
Query 27 RTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKNGFQIG 86
R ++GGNWK NG S+ EL LN A + +T+VV ++++ A+ KL +
Sbjct 4 RKFFVGGNWKMNGDKESLGELIMTLNTASLN-DKTDVVCGAPSIYLDYARSKLDQRIGVA 62
Query 87 CQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMKEEGI 146
QN K GAFTGEI+ M+KD G+ WV++GHSERR + E+D ++ +KVA + E +
Sbjct 63 AQNCYKVPKGAFTGEISPAMIKDCGIDWVILGHSERRHVFGESDELIGQKVAHCL-ESDL 121
Query 147 HVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVATPEQA 206
V CIGEKLEEREA T +V +Q + + V DW RVV+AYEPVWAIGTGK A+PEQA
Sbjct 122 GVIACIGEKLEEREAGTTEDVVFEQTKVIADNVKDWTRVVLAYEPVWAIGTGKTASPEQA 181
Query 207 QEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLKKAFV 266
QEVH LR+++ +AVA+ +RI+YGGSV+ NC+ L DVDGFLVGGASLK FV
Sbjct 182 QEVHEKLREWLRANVSDAVADSVRIIYGGSVTGGNCKELAAQADVDGFLVGGASLKPEFV 241
Query 267 DIIGA 271
DII A
Sbjct 242 DIINA 246
> mmu:21991 Tpi1, AI255506, Tpi, Tpi-1; triosephosphate isomerase
1 (EC:5.3.1.1); K01803 triosephosphate isomerase (TIM) [EC:5.3.1.1]
Length=299
Score = 241 bits (616), Expect = 2e-63, Method: Compositional matrix adjust.
Identities = 128/264 (48%), Positives = 167/264 (63%), Gaps = 5/264 (1%)
Query 8 LCSSAEPHIPFALAAAKMPRTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFP 67
+C+ + P +A R ++GGNWK NG + EL LN A P+ TEVV P
Sbjct 39 VCTDLQRLEPGTMAPT---RKFFVGGNWKMNGRKKCLGELICTLNAANV-PAGTEVVCAP 94
Query 68 TALHIGLAKEKLKNGFQIGCQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYS 127
+I A++KL + QN K GAFTGEI+ GM+KD G WV++GHSERR +
Sbjct 95 PTAYIDFARQKLDPKIAVAAQNCYKVTNGAFTGEISPGMIKDLGATWVVLGHSERRHVFG 154
Query 128 ETDSVVAEKVAMAMKEEGIHVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVI 187
E+D ++ +KV+ A+ E G+ V CIGEKL+EREA T +V +Q + + V DW +VV+
Sbjct 155 ESDELIGQKVSHALAE-GLGVIACIGEKLDEREAGITEKVVFEQTKVIADNVKDWSKVVL 213
Query 188 AYEPVWAIGTGKVATPEQAQEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLR 247
AYEPVWAIGTGK ATP+QAQEVH LR ++ + VA+ RI+YGGSV+ C+ L
Sbjct 214 AYEPVWAIGTGKTATPQQAQEVHEKLRGWLKSNVNDGVAQSTRIIYGGSVTGATCKELAS 273
Query 248 CTDVDGFLVGGASLKKAFVDIIGA 271
DVDGFLVGGASLK FVDII A
Sbjct 274 QPDVDGFLVGGASLKPEFVDIINA 297
> ath:AT3G55440 TPI; TPI (TRIOSEPHOSPHATE ISOMERASE); triose-phosphate
isomerase (EC:5.3.1.1); K01803 triosephosphate isomerase
(TIM) [EC:5.3.1.1]
Length=254
Score = 236 bits (601), Expect = 9e-62, Method: Compositional matrix adjust.
Identities = 125/249 (50%), Positives = 164/249 (65%), Gaps = 4/249 (1%)
Query 25 MPRTPWIGGNWKSNGTVASITELSKELNHAEFDPSQ--TEVVVFPTALHIGLAKEKLKNG 82
M R ++GGNWK NGT + ++ LN A+ PSQ EVVV P + + L K L++
Sbjct 1 MARKFFVGGNWKCNGTAEEVKKIVNTLNEAQV-PSQDVVEVVVSPPYVFLPLVKSTLRSD 59
Query 83 FQIGCQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMK 142
F + QN GAFTGE++A M+ + + WV++GHSERR +E+ V +KVA A+
Sbjct 60 FFVAAQNCWVKKGGAFTGEVSAEMLVNLDIPWVILGHSERRAILNESSEFVGDKVAYALA 119
Query 143 EEGIHVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVAT 202
+ G+ V C+GE LEEREA TM+V Q +A+ +V++W VVIAYEPVWAIGTGKVA+
Sbjct 120 Q-GLKVIACVGETLEEREAGSTMDVVAAQTKAIADRVTNWSNVVIAYEPVWAIGTGKVAS 178
Query 203 PEQAQEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLK 262
P QAQEVH LRK++A+ VA RI+YGGSV+ NC+ L DVDGFLVGGASLK
Sbjct 179 PAQAQEVHDELRKWLAKNVSADVAATTRIIYGGSVNGGNCKELGGQADVDGFLVGGASLK 238
Query 263 KAFVDIIGA 271
F+DII A
Sbjct 239 PEFIDIIKA 247
> tgo:TGME49_033500 triosephosphate isomerase, putative (EC:5.3.1.1);
K01803 triosephosphate isomerase (TIM) [EC:5.3.1.1]
Length=375
Score = 216 bits (550), Expect = 8e-56, Method: Compositional matrix adjust.
Identities = 120/254 (47%), Positives = 159/254 (62%), Gaps = 10/254 (3%)
Query 27 RTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKNG-FQI 85
R ++GGNWK NGT A EL LN A Q +VVV P +L I ++ L+ Q+
Sbjct 118 RRGFVGGNWKCNGTTAKTQELVDMLNSAPVSFEQVDVVVAPPSLFISQVQDSLRQPRVQV 177
Query 86 GCQNIS-KTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYS----ETDSVVAEKVAMA 140
Q+ S + GAFTGE++ M+K+ + WV++GHSERR + E++ VVA+KV A
Sbjct 178 AAQDSSTQQAYGAFTGELSPKMIKEKNIPWVVLGHSERRAGFGGQPGESNQVVAKKVRAA 237
Query 141 MKEEGIHVTICIGEKLEEREANKTMEVCRQQLEAV---LPKVSDWRRVVIAYEPVWAIGT 197
+ E G+ V +CIGE LEERE+ +T +V +QLEAV +P+ W+ +VIAYEPVWAIGT
Sbjct 238 LNE-GLSVILCIGETLEERESGQTQKVLSEQLEAVRQAVPEADAWKSIVIAYEPVWAIGT 296
Query 198 GKVATPEQAQEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVG 257
GK AT AQE H +R ++A+ VAE R++YGGSV N + L DVDGFLVG
Sbjct 297 GKTATAALAQETHRDIRNWLAQAVSPKVAEATRVIYGGSVKGSNAKELFEGEDVDGFLVG 356
Query 258 GASLKKAFVDIIGA 271
GASL FV II A
Sbjct 357 GASLTGDFVSIIDA 370
> ath:AT2G21170 TIM; TIM (TRIOSEPHOSPHATE ISOMERASE); catalytic/
triose-phosphate isomerase (EC:5.3.1.1); K01803 triosephosphate
isomerase (TIM) [EC:5.3.1.1]
Length=306
Score = 213 bits (543), Expect = 4e-55, Method: Compositional matrix adjust.
Identities = 114/225 (50%), Positives = 145/225 (64%), Gaps = 2/225 (0%)
Query 38 NGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKNGFQIGCQNISKTGPGA 97
NGT SI +L +LN A + + +VVV P ++I K L + I QN GA
Sbjct 66 NGTKDSIAKLISDLNSATLE-ADVDVVVSPPFVYIDQVKSSLTDRIDISGQNSWVGKGGA 124
Query 98 FTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMKEEGIHVTICIGEKLE 157
FTGEI+ +KD G KWV++GHSERR E D + +K A A+ EG+ V CIGEKLE
Sbjct 125 FTGEISVEQLKDLGCKWVILGHSERRHVIGEKDEFIGKKAAYAL-SEGLGVIACIGEKLE 183
Query 158 EREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVATPEQAQEVHAALRKFV 217
EREA KT +VC QL+A V W +V+AYEPVWAIGTGKVA+P+QAQEVH A+R ++
Sbjct 184 EREAGKTFDVCFAQLKAFADAVPSWDNIVVAYEPVWAIGTGKVASPQQAQEVHVAVRGWL 243
Query 218 AEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLK 262
+ E VA + RI+YGGSV+ N L + D+DGFLVGGASLK
Sbjct 244 KKNVSEEVASKTRIIYGGSVNGGNSAELAKEEDIDGFLVGGASLK 288
> eco:b3919 tpiA, ECK3911, JW3890, tpi; triosephosphate isomerase
(EC:5.3.1.1); K01803 triosephosphate isomerase (TIM) [EC:5.3.1.1]
Length=255
Score = 197 bits (501), Expect = 3e-50, Method: Compositional matrix adjust.
Identities = 111/249 (44%), Positives = 153/249 (61%), Gaps = 6/249 (2%)
Query 27 RTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKNG-FQI 85
R P + GNWK NG+ + EL L + V + P ++I +AK + + +
Sbjct 2 RHPLVMGNWKLNGSRHMVHELVSNLRKELAGVAGCAVAIAPPEMYIDMAKREAEGSHIML 61
Query 86 GCQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMKEEG 145
G QN+ GAFTGE +A M+KD G +++++GHSERR Y+ E+D ++A+K A+ +KE+G
Sbjct 62 GAQNVDLNLSGAFTGETSAAMLKDIGAQYIIIGHSERRTYHKESDELIAKKFAV-LKEQG 120
Query 146 IHVTICIGEKLEEREANKTMEVCRQQLEAVLPK--VSDWRRVVIAYEPVWAIGTGKVATP 203
+ +CIGE E EA KT EVC +Q++AVL + + VIAYEPVWAIGTGK ATP
Sbjct 121 LTPVLCIGETEAENEAGKTEEVCARQIDAVLKTQGAAAFEGAVIAYEPVWAIGTGKSATP 180
Query 204 EQAQEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLK- 262
QAQ VH +R +A K +AE++ I YGGSV+ N L D+DG LVGGASLK
Sbjct 181 AQAQAVHKFIRDHIA-KVDANIAEQVIIQYGGSVNASNAAELFAQPDIDGALVGGASLKA 239
Query 263 KAFVDIIGA 271
AF I+ A
Sbjct 240 DAFAVIVKA 248
> pfa:PFC0831w triosephophate isomerase, putative (EC:5.3.1.1);
K01803 triosephosphate isomerase (TIM) [EC:5.3.1.1]
Length=357
Score = 125 bits (315), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 81/250 (32%), Positives = 144/250 (57%), Gaps = 19/250 (7%)
Query 33 GNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLK-NGFQI-GC-QN 89
NWK + +L L ++ + +V++ L+I +K+K N +I C Q+
Sbjct 109 ANWKCYLSKEEAYKLIDSLTRIKY-SNYVDVILSLNLLYIPYLLQKIKENNSKIYACSQD 167
Query 90 ISK-TGPGAFTGEITAGMVKDFGLKWVMVGHSERRQ-YYSETDSV--VAEKVAMAMKEEG 145
+S G G FTGE TA +++DFG ++ ++GHSERRQ +Y +++ + KV A+ +
Sbjct 168 VSLVNGFGPFTGETTAKLIQDFGNEYTLIGHSERRQGFYKNGETIEEIVLKVYNAINSK- 226
Query 146 IHVTICIGEKLEE----REANKTMEVCRQQLEAVLPKVS--DWRRVVIAYEPVWAIGTGK 199
+ V +C G+ + + K ME+ R + K+S + + ++IA+EP +AIGTG+
Sbjct 227 LKVILCTGDDYKNCDNPSSSYKMMELLR----LIKTKISKDEMKNIIIAFEPRFAIGTGQ 282
Query 200 VATPEQAQEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGA 259
+ + + + L++ +A++ ++ +EE+ IVYGGS+S+ N + + T VDGFL+G A
Sbjct 283 PVSYDILNKYYYELKRDIAKEIDKSTSEEMMIVYGGSISKSNMKHYVDNTHVDGFLIGKA 342
Query 260 SLKKAFVDII 269
SL + F+DII
Sbjct 343 SLNEDFIDII 352
> tpv:TP04_0676 triosephosphate isomerase (EC:5.3.1.1); K01803
triosephosphate isomerase (TIM) [EC:5.3.1.1]
Length=259
Score = 67.0 bits (162), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 32/87 (36%), Positives = 55/87 (63%), Gaps = 5/87 (5%)
Query 183 RRVVIAYEPVWAIGTGKVATPEQAQEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNC 242
+R+V+AYEP ++IGT K A PE +V L+K + +K + ++ +YGGSV+E+N
Sbjct 168 QRLVVAYEPYYSIGTDKPADPELVNKVIEDLKKSLGDKV-----KGVKFIYGGSVNEENA 222
Query 243 EALLRCTDVDGFLVGGASLKKAFVDII 269
++ ++DG +VG + + FV+II
Sbjct 223 HRYIKKRNIDGIMVGRVAHNEKFVEII 249
Score = 39.7 bits (91), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 32/95 (33%), Positives = 46/95 (48%), Gaps = 9/95 (9%)
Query 34 NWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKNGFQIGC----QN 89
NWK E+ +F P EV+ P+++H+ + K GF C QN
Sbjct 78 NWKCYLNTKLSNEMINLYGSLKF-PENIEVITSPSSIHLASMVSQYK-GFGSRCKPCSQN 135
Query 90 ISKTGP--GAFTGEITAGMVKDFGLKWVMVGHSER 122
+S G +TGEITAGM+K+F LK V + +R
Sbjct 136 VSHYSKSFGPYTGEITAGMLKEF-LKDVNLTGEQR 169
> cel:C25B8.1 kqt-1; potassium channel, KvQLT family member (kqt-1);
K05324 potassium voltage-gated channel KQT-like subfamily,
invertebrate
Length=740
Score = 32.0 bits (71), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 6 GDLCSSAEPHIPFALAAAKMPRTPWIGGNWKSNGTVA 42
GDL S P P + A+MP P + GNW + +V+
Sbjct 12 GDLLSPESPMTPDGMILARMPWHPGLIGNWNTVKSVS 48
> tgo:TGME49_071730 seryl-tRNA synthetase, putative (EC:6.1.1.11)
Length=288
Score = 30.8 bits (68), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 29/52 (55%), Gaps = 5/52 (9%)
Query 182 WRRVVIAYEPVWAIGTGKVATPEQAQEVHAALRKFVAEKAGEAVAEELRIVY 233
W R ++ + + + + TPEQA+E HA LRK G+ + +EL+I +
Sbjct 97 WNRGLLRQQVFEKVESVVICTPEQAEEEHARLRKI-----GKTLLKELQIPF 143
> mmu:108011 Ap4e1, 2310033A20Rik, 9930028M04Rik, AV087807; adaptor-related
protein complex AP-4, epsilon 1; K12400 AP-4 complex
subunit epsilon-1
Length=1122
Score = 30.8 bits (68), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 21/90 (23%), Positives = 40/90 (44%), Gaps = 14/90 (15%)
Query 156 LEEREANKTMEVC-----------RQQLEAVLPKVSD---WRRVVIAYEPVWAIGTGKVA 201
+++ ++ +EVC R+ + AVLP + D + +I + V A+ +
Sbjct 138 VKDLQSTNLVEVCMALTVVSQIFPREMIPAVLPLIEDKLQHSKEIIRRKAVLALYKFYLI 197
Query 202 TPEQAQEVHAALRKFVAEKAGEAVAEELRI 231
P Q Q +H RK + ++ +A L I
Sbjct 198 APNQVQHIHTKFRKALCDRDVGVMAASLHI 227
Lambda K H
0.317 0.133 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10465222448
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40