bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1376_orf1
Length=190
Score E
Sequences producing significant alignments: (Bits) Value
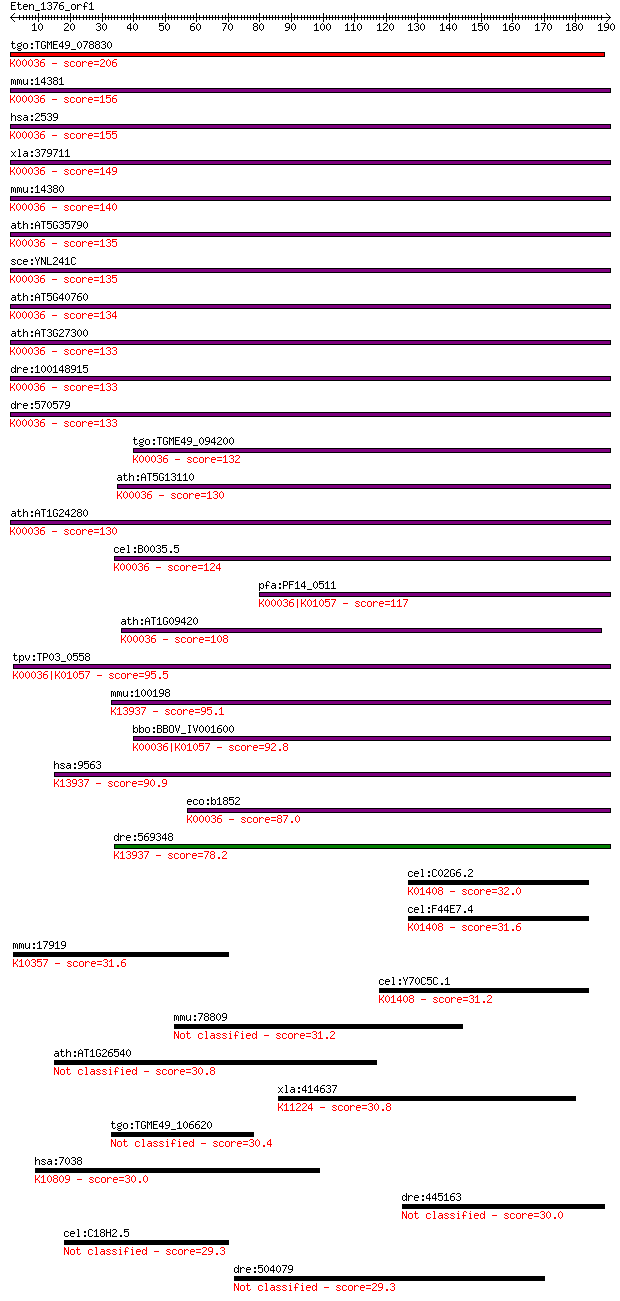
tgo:TGME49_078830 glucose-6-phosphate dehydrogenase, putative ... 206 5e-53
mmu:14381 G6pdx, G28A, G6pd, Gpdx; glucose-6-phosphate dehydro... 156 4e-38
hsa:2539 G6PD, G6PD1; glucose-6-phosphate dehydrogenase (EC:1.... 155 1e-37
xla:379711 g6pd, MGC69058, g6pdh; glucose-6-phosphate dehydrog... 149 5e-36
mmu:14380 G6pd2, G6pdx-ps1, Gpd-2, Gpd2; glucose-6-phosphate d... 140 3e-33
ath:AT5G35790 G6PD1; G6PD1 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE ... 135 1e-31
sce:YNL241C ZWF1, MET19, POS10; Glucose-6-phosphate dehydrogen... 135 1e-31
ath:AT5G40760 G6PD6; G6PD6 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE ... 134 3e-31
ath:AT3G27300 G6PD5; G6PD5 (glucose-6-phosphate dehydrogenase ... 133 3e-31
dre:100148915 glucose-6-phosphate dehydrogenase-like; K00036 g... 133 4e-31
dre:570579 g6pd, fj78b06, si:dkey-90a13.8, wu:fj78b06; glucose... 133 5e-31
tgo:TGME49_094200 glucose-6-phosphate dehydrogenase (EC:1.1.1.... 132 7e-31
ath:AT5G13110 G6PD2; G6PD2 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE ... 130 3e-30
ath:AT1G24280 G6PD3; G6PD3 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE ... 130 3e-30
cel:B0035.5 glucose-6-phosphate-1-dehydrogenase; K00036 glucos... 124 1e-28
pfa:PF14_0511 glucose-6-phosphate dehydrogenase-6-phosphogluco... 117 2e-26
ath:AT1G09420 G6PD4; G6PD4 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE ... 108 2e-23
tpv:TP03_0558 glucose-6-phosphate dehydrogenase-6-phosphogluco... 95.5 9e-20
mmu:100198 H6pd, AI785303, G6pd1, Gpd-1, Gpd1; hexose-6-phosph... 95.1 1e-19
bbo:BBOV_IV001600 21.m02735; glucose-6-phosphate dehydrogenase... 92.8 7e-19
hsa:9563 H6PD, DKFZp686A01246, G6PDH, GDH, MGC87643; hexose-6-... 90.9 3e-18
eco:b1852 zwf, ECK1853, JW1841; glucose-6-phosphate 1-dehydrog... 87.0 4e-17
dre:569348 glucose-6-phosphate dehydrogenase X-linked-like; K1... 78.2 2e-14
cel:C02G6.2 hypothetical protein; K01408 insulysin [EC:3.4.24.56] 32.0 1.5
cel:F44E7.4 hypothetical protein; K01408 insulysin [EC:3.4.24.56] 31.6 1.6
mmu:17919 Myo5b, AI661750, mKIAA1119; myosin VB; K10357 myosin V 31.6 1.8
cel:Y70C5C.1 hypothetical protein; K01408 insulysin [EC:3.4.24... 31.2 2.4
mmu:78809 4930562C15Rik; RIKEN cDNA 4930562C15 gene 31.2 2.5
ath:AT1G26540 agenet domain-containing protein 30.8 2.6
xla:414637 stat5b, MGC81286, stat5; signal transducer and acti... 30.8 3.2
tgo:TGME49_106620 platelet binding protein GspB, putative 30.4 4.2
hsa:7038 TG, AITD3, TGN; thyroglobulin; K10809 thyroglobulin 30.0 4.4
dre:445163 zgc:101119 30.0 4.8
cel:C18H2.5 hypothetical protein 29.3 7.4
dre:504079 im:7151855; si:ch211-253b1.6 29.3 8.5
> tgo:TGME49_078830 glucose-6-phosphate dehydrogenase, putative
(EC:3.1.1.31 1.1.1.49); K00036 glucose-6-phosphate 1-dehydrogenase
[EC:1.1.1.49]
Length=878
Score = 206 bits (523), Expect = 5e-53, Method: Compositional matrix adjust.
Identities = 102/188 (54%), Positives = 132/188 (70%), Gaps = 6/188 (3%)
Query 1 FARSKLDVEEFWKQIAEHLKELSTYFSRQMAPLSSKCDDLVERFRRICTYISGDGYDDAV 60
+ARSK+ ++FW++I++ LK LS++F R+ + + DL+ F+ C+Y+ G YD
Sbjct 417 YARSKMTFDQFWEKISQKLKSLSSFFCRRASAI-----DLLASFKSHCSYLQG-LYDRPA 470
Query 61 ALKKLSNHLDKLEGPEKNNNRLFYLALPPQLFALSVRSIRKHCWAQKGWNRVVVEKPFGR 120
L NHL ++EG + R+ YLALPP +F SV+S R+ CW KGWNRVVVEKPFGR
Sbjct 471 DFANLGNHLKEVEGDAEQVGRVLYLALPPDVFLPSVKSYRQSCWNTKGWNRVVVEKPFGR 530
Query 121 DSESSEKLSNDLMEVLNESEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRI 180
D +SS+KLS LM +L E EI+RIDHYLGKEM+LS+ ALRFANVAF LFHR V VRI
Sbjct 531 DLKSSDKLSASLMALLREREIFRIDHYLGKEMSLSLTALRFANVAFMPLFHRDYVHSVRI 590
Query 181 TFKEDIGT 188
TFKE GT
Sbjct 591 TFKEQSGT 598
> mmu:14381 G6pdx, G28A, G6pd, Gpdx; glucose-6-phosphate dehydrogenase
X-linked (EC:1.1.1.49); K00036 glucose-6-phosphate
1-dehydrogenase [EC:1.1.1.49]
Length=515
Score = 156 bits (395), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 83/198 (41%), Positives = 122/198 (61%), Gaps = 30/198 (15%)
Query 1 FARSKLDV-------EEFWKQIAEHLKELSTYFSRQMAPLSSKCDDLVERFRRICTYISG 53
+ARS+L V E F+K E +L +F+R +Y++G
Sbjct 70 YARSRLTVDDIRKQSEPFFKATPEERPKLEEFFARN-------------------SYVAG 110
Query 54 DGYDDAVALKKLSNHLDKL-EGPEKNNNRLFYLALPPQLFALSVRSIRKHCWAQKGWNRV 112
YDDA + K L++H++ L +G + N RLFYLALPP ++ ++I++ C +Q GWNR+
Sbjct 111 Q-YDDAASYKHLNSHMNALHQGMQAN--RLFYLALPPTVYEAVTKNIQETCMSQTGWNRI 167
Query 113 VVEKPFGRDSESSEKLSNDLMEVLNESEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHR 172
+VEKPFGRD +SS +LSN + + E +IYRIDHYLGKEM +++ LRFAN F +++R
Sbjct 168 IVEKPFGRDLQSSNQLSNHISSLFREDQIYRIDHYLGKEMVQNLMVLRFANRIFGPIWNR 227
Query 173 HNVRCVRITFKEDIGTKG 190
N+ CV +TFKE GT+G
Sbjct 228 DNIACVILTFKEPFGTEG 245
> hsa:2539 G6PD, G6PD1; glucose-6-phosphate dehydrogenase (EC:1.1.1.49);
K00036 glucose-6-phosphate 1-dehydrogenase [EC:1.1.1.49]
Length=545
Score = 155 bits (391), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 81/197 (41%), Positives = 121/197 (61%), Gaps = 28/197 (14%)
Query 1 FARSKLDV-------EEFWKQIAEHLKELSTYFSRQMAPLSSKCDDLVERFRRICTYISG 53
+ARS+L V E F+K E +L +F+R +Y++G
Sbjct 100 YARSRLTVADIRKQSEPFFKATPEEKLKLEDFFARN-------------------SYVAG 140
Query 54 DGYDDAVALKKLSNHLDKLEGPEKNNNRLFYLALPPQLFALSVRSIRKHCWAQKGWNRVV 113
YDDA + ++L++H++ L + N RLFYLALPP ++ ++I + C +Q GWNR++
Sbjct 141 Q-YDDAASYQRLNSHMNALHLGSQAN-RLFYLALPPTVYEAVTKNIHESCMSQIGWNRII 198
Query 114 VEKPFGRDSESSEKLSNDLMEVLNESEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRH 173
VEKPFGRD +SS++LSN + + E +IYRIDHYLGKEM +++ LRFAN F +++R
Sbjct 199 VEKPFGRDLQSSDRLSNHISSLFREDQIYRIDHYLGKEMVQNLMVLRFANRIFGPIWNRD 258
Query 174 NVRCVRITFKEDIGTKG 190
N+ CV +TFKE GT+G
Sbjct 259 NIACVILTFKEPFGTEG 275
> xla:379711 g6pd, MGC69058, g6pdh; glucose-6-phosphate dehydrogenase
(EC:1.1.1.49); K00036 glucose-6-phosphate 1-dehydrogenase
[EC:1.1.1.49]
Length=518
Score = 149 bits (377), Expect = 5e-36, Method: Compositional matrix adjust.
Identities = 84/197 (42%), Positives = 115/197 (58%), Gaps = 28/197 (14%)
Query 1 FARSKLDVEEFWKQIAEHLK-------ELSTYFSRQMAPLSSKCDDLVERFRRICTYISG 53
FARSKL V++ KQ + K +L T+F R +YISG
Sbjct 73 FARSKLTVQDIKKQSEPYFKVSAEDALKLDTFFKRN-------------------SYISG 113
Query 54 DGYDDAVALKKLSNHLDKLEGPEKNNNRLFYLALPPQLFALSVRSIRKHCWAQKGWNRVV 113
Y DA + + L+ HL+ L K N RLFYLALPP ++ R+I++ C + GWNRV+
Sbjct 114 Q-YSDAASFQNLNQHLNSLPNGAKAN-RLFYLALPPSVYHDVTRNIKETCMSSVGWNRVI 171
Query 114 VEKPFGRDSESSEKLSNDLMEVLNESEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRH 173
VEKPFG+D ESS +LS + + E++IYRIDHYLGKEM +++ LRF N F L+ R
Sbjct 172 VEKPFGKDLESSNRLSEHISSLYKENQIYRIDHYLGKEMVQNLMILRFGNRIFSPLWSRD 231
Query 174 NVRCVRITFKEDIGTKG 190
++ V +TFKE GT+G
Sbjct 232 HISAVVLTFKEPFGTQG 248
> mmu:14380 G6pd2, G6pdx-ps1, Gpd-2, Gpd2; glucose-6-phosphate
dehydrogenase 2 (EC:1.1.1.49); K00036 glucose-6-phosphate 1-dehydrogenase
[EC:1.1.1.49]
Length=513
Score = 140 bits (352), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 77/198 (38%), Positives = 118/198 (59%), Gaps = 30/198 (15%)
Query 1 FARSKLDV-------EEFWKQIAEHLKELSTYFSRQMAPLSSKCDDLVERFRRICTYISG 53
+ARS+L V E F+K E +L +F+R +Y+ G
Sbjct 70 YARSQLTVDDIQKQSEPFFKATPEERPKLEEFFTRN-------------------SYVVG 110
Query 54 DGYDDAVALKKLSNHLDKL-EGPEKNNNRLFYLALPPQLFALSVRSIRKHCWAQKGWNRV 112
YDD + K L+++++ L +G + N+ LFYLALPP ++ ++I++ C +Q G+NR+
Sbjct 111 Q-YDDPASYKHLNSYINALHQGMQANH--LFYLALPPTVYEAVTKNIQETCMSQTGFNRI 167
Query 113 VVEKPFGRDSESSEKLSNDLMEVLNESEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHR 172
+VEKPFGRD +SS +LSN + + E +IYRIDHYL KEM +++ LRFAN F +++
Sbjct 168 IVEKPFGRDLQSSNQLSNHISSLFREDQIYRIDHYLDKEMVQNLMVLRFANRIFGPIWNG 227
Query 173 HNVRCVRITFKEDIGTKG 190
N+ CV +TFKE GT+G
Sbjct 228 DNIACVILTFKEPFGTEG 245
> ath:AT5G35790 G6PD1; G6PD1 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE
1); glucose-6-phosphate dehydrogenase (EC:1.1.1.49); K00036
glucose-6-phosphate 1-dehydrogenase [EC:1.1.1.49]
Length=576
Score = 135 bits (340), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 79/190 (41%), Positives = 114/190 (60%), Gaps = 10/190 (5%)
Query 1 FARSKLDVEEFWKQIAEHLKELSTYFSRQMAPLSSKCDDLVERFRRICTYISGDGYDDAV 60
+AR+KL EE I+ ST R KC D +E+F + C Y SG Y+
Sbjct 129 YARTKLTHEELRDMIS------STLTCR--IDQREKCGDKMEQFLKRCFYHSGQ-YNSEE 179
Query 61 ALKKLSNHLDKLEGPEKNNNRLFYLALPPQLFALSVRSIRKHCWAQKGWNRVVVEKPFGR 120
+L+ L + E K +NRL+YL++PP +F VR ++ GW RV+VEKPFGR
Sbjct 180 DFAELNKKLKEKEA-GKISNRLYYLSIPPNIFVDVVRCASLRASSENGWTRVIVEKPFGR 238
Query 121 DSESSEKLSNDLMEVLNESEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRI 180
DSESS +L+ L + L E +I+RIDHYLGKE+ ++ LRF+N+ F+ L+ R+ +R V++
Sbjct 239 DSESSGELTRCLKQYLTEEQIFRIDHYLGKELVENLSVLRFSNLVFEPLWSRNYIRNVQL 298
Query 181 TFKEDIGTKG 190
F ED GT+G
Sbjct 299 IFSEDFGTEG 308
> sce:YNL241C ZWF1, MET19, POS10; Glucose-6-phosphate dehydrogenase
(G6PD), catalyzes the first step of the pentose phosphate
pathway; involved in adapting to oxidatve stress; homolog
of the human G6PD which is deficient in patients with hemolytic
anemia (EC:1.1.1.49); K00036 glucose-6-phosphate 1-dehydrogenase
[EC:1.1.1.49]
Length=505
Score = 135 bits (339), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 76/193 (39%), Positives = 113/193 (58%), Gaps = 14/193 (7%)
Query 1 FARSKLDVEEFWK-QIAEHLKELSTYFSRQMAPLSSKCDDLVERFRRICTYISGDGYDDA 59
+ARSKL +EE K ++ HLK+ P D VE+F ++ +YISG+ YD
Sbjct 50 YARSKLSMEEDLKSRVLPHLKK----------PHGEADDSKVEQFFKMVSYISGN-YDTD 98
Query 60 VALKKLSNHLDKLEGPEKNN--NRLFYLALPPQLFALSVRSIRKHCWAQKGWNRVVVEKP 117
+L ++K E + +RLFYLALPP +F + I+ +A+ G RV+VEKP
Sbjct 99 EGFDELRTQIEKFEKSANVDVPHRLFYLALPPSVFLTVAKQIKSRVYAENGITRVIVEKP 158
Query 118 FGRDSESSEKLSNDLMEVLNESEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRC 177
FG D S+ +L +L + E E+YRIDHYLGKE+ +++ LRF N ++R N++
Sbjct 159 FGHDLASARELQKNLGPLFKEEELYRIDHYLGKELVKNLLVLRFGNQFLNASWNRDNIQS 218
Query 178 VRITFKEDIGTKG 190
V+I+FKE GT+G
Sbjct 219 VQISFKERFGTEG 231
> ath:AT5G40760 G6PD6; G6PD6 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE
6); glucose-6-phosphate dehydrogenase; K00036 glucose-6-phosphate
1-dehydrogenase [EC:1.1.1.49]
Length=515
Score = 134 bits (336), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 77/197 (39%), Positives = 118/197 (59%), Gaps = 18/197 (9%)
Query 1 FARSKLDVEEFWKQIAEHLKELSTYFSRQMAPLSSKCDDLVERFRRICTYISGDGYDDAV 60
+AR+K+ EE +I +L + + Q LS +F ++ Y+SG YD
Sbjct 71 YARTKISDEELRDRIRGYLVDEKN--AEQAEALS--------KFLQLIKYVSGP-YDAEE 119
Query 61 ALKKLSNHLDKLE----GPEKNNNRLFYLALPPQLFALSVRSIRKHCWAQK---GWNRVV 113
++L + + E E ++ RLFYLALPP ++ + I+ C + GW R+V
Sbjct 120 GFQRLDKAISEHEISKNSTEGSSRRLFYLALPPSVYPSVCKMIKTCCMNKSDLGGWTRIV 179
Query 114 VEKPFGRDSESSEKLSNDLMEVLNESEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRH 173
VEKPFG+D ES+E+LS+ + E+ +ES+IYRIDHYLGKE+ +++ LRFAN F L++R
Sbjct 180 VEKPFGKDLESAEQLSSQIGELFDESQIYRIDHYLGKELVQNMLVLRFANRFFLPLWNRD 239
Query 174 NVRCVRITFKEDIGTKG 190
N+ V+I F+ED GT+G
Sbjct 240 NIENVQIVFREDFGTEG 256
> ath:AT3G27300 G6PD5; G6PD5 (glucose-6-phosphate dehydrogenase
5); glucose-6-phosphate dehydrogenase (EC:1.1.1.49); K00036
glucose-6-phosphate 1-dehydrogenase [EC:1.1.1.49]
Length=516
Score = 133 bits (335), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 76/197 (38%), Positives = 116/197 (58%), Gaps = 17/197 (8%)
Query 1 FARSKLDVEEFWKQIAEHLKELSTYFSRQMAPLSSKCDDLVERFRRICTYISGDGYDDAV 60
+ARSK+ EE +I +L + +SK + + +F ++ Y+SG YD
Sbjct 71 YARSKITDEELRDKIRGYLVDEKN---------ASKKTEALSKFLKLIKYVSGP-YDSEE 120
Query 61 ALKKLSNHLDKLE----GPEKNNNRLFYLALPPQLFALSVRSIRKHCWAQK---GWNRVV 113
K+L + + E E ++ RLFYLALPP ++ + I+ C + GW R+V
Sbjct 121 GFKRLDKAILEHEISKKTAEGSSRRLFYLALPPSVYPPVSKMIKAWCTNKSDLGGWTRIV 180
Query 114 VEKPFGRDSESSEKLSNDLMEVLNESEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRH 173
VEKPFG+D ES+E+LS+ + + E +IYRIDHYLGKE+ +++ LRFAN F L++R
Sbjct 181 VEKPFGKDLESAEQLSSQIGALFEEPQIYRIDHYLGKELVQNMLVLRFANRLFLPLWNRD 240
Query 174 NVRCVRITFKEDIGTKG 190
N+ V+I F+ED GT+G
Sbjct 241 NIANVQIVFREDFGTEG 257
> dre:100148915 glucose-6-phosphate dehydrogenase-like; K00036
glucose-6-phosphate 1-dehydrogenase [EC:1.1.1.49]
Length=523
Score = 133 bits (334), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 82/197 (41%), Positives = 113/197 (57%), Gaps = 28/197 (14%)
Query 1 FARSKLDVE-------EFWKQIAEHLKELSTYFSRQMAPLSSKCDDLVERFRRICTYISG 53
FARS L V+ + K + + L+ +FSR +YISG
Sbjct 78 FARSDLTVDAIRIACMPYMKVVDNEAERLAAFFSRN-------------------SYISG 118
Query 54 DGYDDAVALKKLSNHLDKLEGPEKNNNRLFYLALPPQLFALSVRSIRKHCWAQKGWNRVV 113
Y + + L+ HL L G NRLFYLALPP ++ ++I+ C + KGWNRV+
Sbjct 119 K-YVEESSFSDLNTHLLSLPG-GAEANRLFYLALPPSVYHDVTKNIKHQCMSTKGWNRVI 176
Query 114 VEKPFGRDSESSEKLSNDLMEVLNESEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRH 173
VEKPFGRD +SSE+LS+ L + E +IYRIDHYLGKEM +++ LRF N F +++R
Sbjct 177 VEKPFGRDLQSSEELSSHLSSLFTEEQIYRIDHYLGKEMVQNLMVLRFGNRIFGPIWNRD 236
Query 174 NVRCVRITFKEDIGTKG 190
+V CV +TFKE GT+G
Sbjct 237 SVACVVLTFKEPFGTQG 253
> dre:570579 g6pd, fj78b06, si:dkey-90a13.8, wu:fj78b06; glucose-6-phosphate
dehydrogenase; K00036 glucose-6-phosphate 1-dehydrogenase
[EC:1.1.1.49]
Length=523
Score = 133 bits (334), Expect = 5e-31, Method: Compositional matrix adjust.
Identities = 82/197 (41%), Positives = 114/197 (57%), Gaps = 28/197 (14%)
Query 1 FARSKLDVE-------EFWKQIAEHLKELSTYFSRQMAPLSSKCDDLVERFRRICTYISG 53
FARS L V+ + K + + L+ +FSR +YISG
Sbjct 78 FARSDLTVDAIRIACMPYMKVVDNEAERLAAFFSRN-------------------SYISG 118
Query 54 DGYDDAVALKKLSNHLDKLEGPEKNNNRLFYLALPPQLFALSVRSIRKHCWAQKGWNRVV 113
Y + + L+ HL L G + N RLFYLALPP ++ ++I+ C + KGWNRV+
Sbjct 119 K-YVEESSFSDLNTHLLSLPGGAEAN-RLFYLALPPSVYHDVTKNIKHQCMSTKGWNRVI 176
Query 114 VEKPFGRDSESSEKLSNDLMEVLNESEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRH 173
VEKPFGRD +SSE+LS+ L + E +IYRIDHYLGKEM +++ LRF N F +++R
Sbjct 177 VEKPFGRDLQSSEELSSHLSSLFTEEQIYRIDHYLGKEMVQNLMVLRFGNRIFGPIWNRD 236
Query 174 NVRCVRITFKEDIGTKG 190
+V CV +TFKE GT+G
Sbjct 237 SVACVVLTFKEPFGTQG 253
> tgo:TGME49_094200 glucose-6-phosphate dehydrogenase (EC:1.1.1.49);
K00036 glucose-6-phosphate 1-dehydrogenase [EC:1.1.1.49]
Length=560
Score = 132 bits (332), Expect = 7e-31, Method: Compositional matrix adjust.
Identities = 68/154 (44%), Positives = 94/154 (61%), Gaps = 4/154 (2%)
Query 40 LVERFRRICTYISGDGYDDAVALKKLSNHLDKLEGPEKNN---NRLFYLALPPQLFALSV 96
L+ +F + +Y +G DD + L +++ ++E + N R+ YLALPP +FA +
Sbjct 128 LLNQFEQRMSYTTGSIDDDNI-LSHFCHNISRMEQAQSPNASWGRVLYLALPPHIFAPAA 186
Query 97 RSIRKHCWAQKGWNRVVVEKPFGRDSESSEKLSNDLMEVLNESEIYRIDHYLGKEMTLSI 156
+++C GW RVVVEKPFGRD ESSE LS L VL E E YRIDHYLGKEM ++
Sbjct 187 AGFKRNCSTHNGWTRVVVEKPFGRDYESSELLSEQLRSVLLEEETYRIDHYLGKEMLQAL 246
Query 157 IALRFANVAFKHLFHRHNVRCVRITFKEDIGTKG 190
LRF N + L +R+ V+ + I+F EDIG G
Sbjct 247 PPLRFTNFFLEPLMNRNFVKALTISFNEDIGISG 280
> ath:AT5G13110 G6PD2; G6PD2 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE
2); glucose-6-phosphate dehydrogenase (EC:1.1.1.49); K00036
glucose-6-phosphate 1-dehydrogenase [EC:1.1.1.49]
Length=596
Score = 130 bits (327), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 66/156 (42%), Positives = 96/156 (61%), Gaps = 2/156 (1%)
Query 35 SKCDDLVERFRRICTYISGDGYDDAVALKKLSNHLDKLEGPEKNNNRLFYLALPPQLFAL 94
+ C + +E F + C Y SG YD +L L + E + +NRLFYL++PP +F
Sbjct 174 ANCGEKMEEFLKRCFYHSGQ-YDSQEHFTELDKKLKEHEA-GRISNRLFYLSIPPNIFVD 231
Query 95 SVRSIRKHCWAQKGWNRVVVEKPFGRDSESSEKLSNDLMEVLNESEIYRIDHYLGKEMTL 154
+V+ + GW RV+VEKPFGRDSE+S L+ L + L E +I+RIDHYLGKE+
Sbjct 232 AVKCASTSASSVNGWTRVIVEKPFGRDSETSAALTKSLKQYLEEDQIFRIDHYLGKELVE 291
Query 155 SIIALRFANVAFKHLFHRHNVRCVRITFKEDIGTKG 190
++ LRF+N+ F+ L+ R +R V+ F ED GT+G
Sbjct 292 NLSVLRFSNLIFEPLWSRQYIRNVQFIFSEDFGTEG 327
> ath:AT1G24280 G6PD3; G6PD3 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE
3); glucose-6-phosphate dehydrogenase (EC:1.1.1.49); K00036
glucose-6-phosphate 1-dehydrogenase [EC:1.1.1.49]
Length=599
Score = 130 bits (326), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 73/190 (38%), Positives = 110/190 (57%), Gaps = 10/190 (5%)
Query 1 FARSKLDVEEFWKQIAEHLKELSTYFSRQMAPLSSKCDDLVERFRRICTYISGDGYDDAV 60
+ARSK+ E ++ K L+ ++ + C + +E F + C Y SG YD
Sbjct 151 YARSKMTDAELRVMVS---KTLTCRIDKR-----ANCGEKMEEFLKRCFYHSGQ-YDSQE 201
Query 61 ALKKLSNHLDKLEGPEKNNNRLFYLALPPQLFALSVRSIRKHCWAQKGWNRVVVEKPFGR 120
L L + EG + +NRLFYL++PP +F +V+ + GW RV+VEKPFGR
Sbjct 202 HFVALDEKLKEHEG-GRLSNRLFYLSIPPNIFVDAVKCASSSASSVNGWTRVIVEKPFGR 260
Query 121 DSESSEKLSNDLMEVLNESEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRI 180
DS++S L+ L + L E +I+RIDHYLGKE+ ++ LRF+N+ F+ L+ R +R V+
Sbjct 261 DSKTSAALTKSLKQYLEEDQIFRIDHYLGKELVENLSVLRFSNLIFEPLWSRQYIRNVQF 320
Query 181 TFKEDIGTKG 190
F ED GT+G
Sbjct 321 IFSEDFGTEG 330
> cel:B0035.5 glucose-6-phosphate-1-dehydrogenase; K00036 glucose-6-phosphate
1-dehydrogenase [EC:1.1.1.49]
Length=522
Score = 124 bits (312), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 64/162 (39%), Positives = 98/162 (60%), Gaps = 10/162 (6%)
Query 34 SSKC--DDLVERFRRICTYISGDGYDDAVALKKLSNHLDKLEGPEKNN--NRLFYLALPP 89
+ KC DD +++ C+Y+ G YD + ++L + +D + N NRL+YLALPP
Sbjct 95 NEKCAFDDFIKK----CSYVQGQ-YDTSEGFQRLQSSIDDFQKESNNQAVNRLYYLALPP 149
Query 90 QLFALSVRSIRKHCWAQ-KGWNRVVVEKPFGRDSESSEKLSNDLMEVLNESEIYRIDHYL 148
+F + ++K+C W RV++EKPFG D +SS +LS L ++ E +IYRIDHYL
Sbjct 150 SVFNVVSTELKKNCMDHGDSWTRVIIEKPFGHDLKSSCELSTHLAKLFKEDQIYRIDHYL 209
Query 149 GKEMTLSIIALRFANVAFKHLFHRHNVRCVRITFKEDIGTKG 190
GKEM +++ +RF N ++R ++ V I+FKED GT G
Sbjct 210 GKEMVQNLMVMRFGNRILAPSWNRDHIASVMISFKEDFGTGG 251
> pfa:PF14_0511 glucose-6-phosphate dehydrogenase-6-phosphogluconolactonase
(EC:1.1.1.49); K00036 glucose-6-phosphate 1-dehydrogenase
[EC:1.1.1.49]; K01057 6-phosphogluconolactonase
[EC:3.1.1.31]
Length=910
Score = 117 bits (293), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 49/111 (44%), Positives = 76/111 (68%), Gaps = 0/111 (0%)
Query 80 NRLFYLALPPQLFALSVRSIRKHCWAQKGWNRVVVEKPFGRDSESSEKLSNDLMEVLNES 139
NR+ YLALPP +F ++++ +K+C KG +++++EKPFG D +S + LS ++E NE
Sbjct 512 NRMLYLALPPHIFVSTLKNYKKNCLNSKGTDKILLEKPFGNDLDSFKMLSKQILENFNEQ 571
Query 140 EIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRITFKEDIGTKG 190
+IYRIDHYLGK+M ++ L+F N L +RH ++C++IT KE G G
Sbjct 572 QIYRIDHYLGKDMVSGLLKLKFTNTFLLSLMNRHFIKCIKITLKETKGVYG 622
> ath:AT1G09420 G6PD4; G6PD4 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE
4); glucose-6-phosphate dehydrogenase; K00036 glucose-6-phosphate
1-dehydrogenase [EC:1.1.1.49]
Length=625
Score = 108 bits (269), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 52/152 (34%), Positives = 91/152 (59%), Gaps = 2/152 (1%)
Query 36 KCDDLVERFRRICTYISGDGYDDAVALKKLSNHLDKLEGPEKNNNRLFYLALPPQLFALS 95
C ++ F+ YI+G GY++ + +L+ + ++EG E NR+FYL++P +
Sbjct 219 NCGGKMDAFQSRTYYING-GYNNRDGMSRLAERMKQIEG-ESEANRIFYLSVPQEALVDV 276
Query 96 VRSIRKHCWAQKGWNRVVVEKPFGRDSESSEKLSNDLMEVLNESEIYRIDHYLGKEMTLS 155
+I + A +GW R++VEKPFG +S SS +L+ L+ E +IYRIDH LG+ + +
Sbjct 277 ACTIGDNAQAPRGWTRIIVEKPFGFNSHSSHQLTKSLLSKFEEKQIYRIDHMLGRNLIEN 336
Query 156 IIALRFANVAFKHLFHRHNVRCVRITFKEDIG 187
+ LRF+N+ F+ L++R +R +++ E I
Sbjct 337 LTVLRFSNLVFEPLWNRTYIRNIQVIISESIA 368
> tpv:TP03_0558 glucose-6-phosphate dehydrogenase-6-phosphogluconolactonase
(EC:1.1.1.49); K00036 glucose-6-phosphate 1-dehydrogenase
[EC:1.1.1.49]; K01057 6-phosphogluconolactonase
[EC:3.1.1.31]
Length=869
Score = 95.5 bits (236), Expect = 9e-20, Method: Compositional matrix adjust.
Identities = 55/190 (28%), Positives = 102/190 (53%), Gaps = 3/190 (1%)
Query 2 ARSKLDVEEFWKQIAEHL-KELSTYFSRQMAPLSSKCDDLVERFRRICTYISGDGYDDAV 60
+RS D EEF+ QI+ + ++T + + ++ F+ C+ I YDD+
Sbjct 423 SRSHCDFEEFFSQISNDIFSSITTNIFMRNPAVRFDFPSVITEFKSRCSRICLR-YDDSS 481
Query 61 ALKKLSNHLDKLEGPEKNNNRLFYLALPPQLFALSVRSIRKHCWAQKGWNRVVVEKPFGR 120
++ L ++E + ++R+ YLA P + + +R + C + GW RV++EKPFGR
Sbjct 482 FFERFKKTLREIEHNSETSHRMVYLATPSEAYQNILRVVTSCCKPENGWFRVMLEKPFGR 541
Query 121 DSESSEKLSNDLMEVLNESEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRI 180
D +S E++ L + + E + +DHYLGK + I++ + + + +F+R ++ V I
Sbjct 542 DLQSCEEIDRFLNKHVAPDEAFLVDHYLGKPVVGCILSTKIGS-NYARIFNRRYIKSVHI 600
Query 181 TFKEDIGTKG 190
KE+IG+ G
Sbjct 601 LLKEEIGSFG 610
> mmu:100198 H6pd, AI785303, G6pd1, Gpd-1, Gpd1; hexose-6-phosphate
dehydrogenase (glucose 1-dehydrogenase) (EC:1.1.1.47 3.1.1.31);
K13937 hexose-6-phosphate dehydrogenase [EC:1.1.1.47
3.1.1.31]
Length=797
Score = 95.1 bits (235), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 57/160 (35%), Positives = 85/160 (53%), Gaps = 3/160 (1%)
Query 33 LSSKCDDLVERFRRICTYISGDGYDDAVALKKLSNHLDKLEGPEKNNNRLFYLALPPQLF 92
+ S+CD+L +F ++ Y +D L K + +G R+FY ++PP +
Sbjct 96 VPSRCDELKGQFLQLSQYRQLKTVEDYQTLNKDIETQVQQDG-LWEAGRIFYFSVPPFAY 154
Query 93 ALSVRSIRKHCWAQKG-WNRVVVEKPFGRDSESSEKLSNDLMEVLNESEIYRIDHYLGKE 151
A R+I C G W RVV EKPFG D S+++L+++L E E+YR+DHYLGK+
Sbjct 155 ADIARNINSSCRPHPGAWLRVVFEKPFGHDHLSAQQLASELGSFFQEEEMYRVDHYLGKQ 214
Query 152 MTLSIIALRFAN-VAFKHLFHRHNVRCVRITFKEDIGTKG 190
I+ R N A L++RH+V V I KE I +G
Sbjct 215 AVAQILPFRDQNRKALDGLWNRHHVERVEIILKETIDAEG 254
> bbo:BBOV_IV001600 21.m02735; glucose-6-phosphate dehydrogenase-6-phosphogluconolactonase
(EC:1.1.1.49); K00036 glucose-6-phosphate
1-dehydrogenase [EC:1.1.1.49]; K01057 6-phosphogluconolactonase
[EC:3.1.1.31]
Length=806
Score = 92.8 bits (229), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 53/151 (35%), Positives = 84/151 (55%), Gaps = 2/151 (1%)
Query 40 LVERFRRICTYISGDGYDDAVALKKLSNHLDKLEGPEKNNNRLFYLALPPQLFALSVRSI 99
++ F+++ I+ YDD + KL+ L +LE +RL YLA P + + ++
Sbjct 449 IISEFKKVLRRITIK-YDDPESETKLNETLKELECGSAVTHRLVYLATPAEAYHPIMKLA 507
Query 100 RKHCWAQKGWNRVVVEKPFGRDSESSEKLSNDLMEVLNESEIYRIDHYLGKEMTLSIIAL 159
C GW RV++EKPFGRD SS+++ L+E E++ +DHYLGK + +IA+
Sbjct 508 TSVCRPANGWFRVILEKPFGRDLGSSQQIQKVLVEHATSDEVFLVDHYLGKPLISCMIAI 567
Query 160 RFANVAFKHLFHRHNVRCVRITFKEDIGTKG 190
R +V + +LF V+ V I KE IG+ G
Sbjct 568 R-RSVRYTNLFCNKYVKSVHIKMKETIGSFG 597
> hsa:9563 H6PD, DKFZp686A01246, G6PDH, GDH, MGC87643; hexose-6-phosphate
dehydrogenase (glucose 1-dehydrogenase) (EC:1.1.1.47
3.1.1.31); K13937 hexose-6-phosphate dehydrogenase [EC:1.1.1.47
3.1.1.31]
Length=791
Score = 90.9 bits (224), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 61/178 (34%), Positives = 93/178 (52%), Gaps = 7/178 (3%)
Query 15 IAEHLKELSTYFSRQMAPLSSKCDDLVERFRRICTYISGDGYDDAVALKKLSNHLDKLEG 74
+A+ L+ LS + MAP S C + ++F ++ Y +D AL K + G
Sbjct 77 MAKALESLSC--PKDMAP--SHCAEHKDQFLQLSQYRQLKTAEDYQALNKDIEAQLQHAG 132
Query 75 PEKNNNRLFYLALPPQLFALSVRSIRKHCWAQKG-WNRVVVEKPFGRDSESSEKLSNDLM 133
+ R+FY ++PP + R+I C G W RVV+EKPFG D S+++L+ +L
Sbjct 133 -LREAGRIFYFSVPPFAYEDIARNINSSCRPGPGAWLRVVLEKPFGHDHFSAQQLATELG 191
Query 134 EVLNESEIYRIDHYLGKEMTLSIIALRFAN-VAFKHLFHRHNVRCVRITFKEDIGTKG 190
E E+YR+DHYLGK+ I+ R N A L++RH+V V I KE + +G
Sbjct 192 TFFQEEEMYRVDHYLGKQAVAQILPFRDQNRKALDGLWNRHHVERVEIIMKETVDAEG 249
> eco:b1852 zwf, ECK1853, JW1841; glucose-6-phosphate 1-dehydrogenase
(EC:1.1.1.49); K00036 glucose-6-phosphate 1-dehydrogenase
[EC:1.1.1.49]
Length=491
Score = 87.0 bits (214), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 47/134 (35%), Positives = 74/134 (55%), Gaps = 6/134 (4%)
Query 57 DDAVALKKLSNHLDKLEGPEKNNNRLFYLALPPQLFALSVRSIRKHCWAQKGWNRVVVEK 116
+D A +L LD +KN + Y A+PP F + + + K RVV+EK
Sbjct 94 NDTAAFSRLGAMLD-----QKNRITINYFAMPPSTFGAICKGLGEAKLNAKP-ARVVMEK 147
Query 117 PFGRDSESSEKLSNDLMEVLNESEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVR 176
P G +S+++++ + E E ++YRIDHYLGKE L+++ALRFAN F + + +
Sbjct 148 PLGTSLATSQEINDQVGEYFEECQVYRIDHYLGKETVLNLLALRFANSLFVNNWDNRTID 207
Query 177 CVRITFKEDIGTKG 190
V IT E++G +G
Sbjct 208 HVEITVAEEVGIEG 221
> dre:569348 glucose-6-phosphate dehydrogenase X-linked-like;
K13937 hexose-6-phosphate dehydrogenase [EC:1.1.1.47 3.1.1.31]
Length=781
Score = 78.2 bits (191), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 52/161 (32%), Positives = 79/161 (49%), Gaps = 7/161 (4%)
Query 34 SSKCDDLVERFRRICTYISGDGYDDAVALKKLSNHLDKLEGPEKNN--NRLFYLALPPQL 91
+ +C + E+F + Y +D +KL + + G E RLFYL++P
Sbjct 89 AERCALVKEQFLHLSRYHQLKTAED---YEKLCQQIKQQVGQESMTEAGRLFYLSVPAFA 145
Query 92 FALSVRSIRKHC-WAQKGWNRVVVEKPFGRDSESSEKLSNDLMEVLNESEIYRIDHYLGK 150
+A I C W RVV+EKPFG D S++ L L L E E+YRIDHYLGK
Sbjct 146 YAEIAERINNTCRPPSDAWLRVVLEKPFGHDFASAQLLDKKLSGQLKEEEMYRIDHYLGK 205
Query 151 EMTLSIIALRFANVA-FKHLFHRHNVRCVRITFKEDIGTKG 190
++ I+ R N ++++H++ + I KE + KG
Sbjct 206 QVVSKILPFRKENKKLLDPIWNKHHIERIEIVLKETLDAKG 246
> cel:C02G6.2 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=816
Score = 32.0 bits (71), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 32/62 (51%), Gaps = 9/62 (14%)
Query 127 KLSNDLMEVL-----NESEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRIT 181
+LSN+LM++L N +YR +H +E L+ ++HL H+ CV +T
Sbjct 722 QLSNELMDILKSAAPNSRLLYRNEHNPRREFQLN----NGDEYIYRHLQKTHDAGCVEVT 777
Query 182 FK 183
FK
Sbjct 778 FK 779
> cel:F44E7.4 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=1051
Score = 31.6 bits (70), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 33/62 (53%), Gaps = 9/62 (14%)
Query 127 KLSNDLMEVL-----NESEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRIT 181
+LS +LM+VL N +YR +H +E+ L+ ++HL H+V CV +T
Sbjct 765 QLSKELMDVLKSAAPNSRPLYRNEHNPRRELQLN----NGDEYVYRHLQKTHDVGCVEVT 820
Query 182 FK 183
++
Sbjct 821 YQ 822
> mmu:17919 Myo5b, AI661750, mKIAA1119; myosin VB; K10357 myosin
V
Length=1818
Score = 31.6 bits (70), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 32/68 (47%), Gaps = 0/68 (0%)
Query 2 ARSKLDVEEFWKQIAEHLKELSTYFSRQMAPLSSKCDDLVERFRRICTYISGDGYDDAVA 61
AR +L + + AEHLK L+ ++ L K DD + F+ + +S AV
Sbjct 900 ARQELKALKIEARSAEHLKRLNVGMENKVVQLQRKIDDQNKEFKTLSEQLSAVTSSHAVE 959
Query 62 LKKLSNHL 69
++KL L
Sbjct 960 VEKLKKEL 967
> cel:Y70C5C.1 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=985
Score = 31.2 bits (69), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 39/72 (54%), Gaps = 10/72 (13%)
Query 118 FGRDSESSE-KLSNDLMEVLNES-----EIYRIDHYLGKEMTLSIIALRFANVAFKHLFH 171
+G +E +LS DL+++L + ++R +H L +E+ L+ ++HL
Sbjct 696 YGNSTEKETIQLSKDLIDILKSAAPSSRPLFRNEHILRREIQLN----NGDEYIYRHLQT 751
Query 172 RHNVRCVRITFK 183
H+V CV++T++
Sbjct 752 THDVGCVQVTYQ 763
> mmu:78809 4930562C15Rik; RIKEN cDNA 4930562C15 gene
Length=1145
Score = 31.2 bits (69), Expect = 2.5, Method: Composition-based stats.
Identities = 22/92 (23%), Positives = 45/92 (48%), Gaps = 1/92 (1%)
Query 53 GDGYDDAVALKKLSNHLDKL-EGPEKNNNRLFYLALPPQLFALSVRSIRKHCWAQKGWNR 111
GD +K+LSN D++ + EK ++L + P L S A++ WN
Sbjct 65 GDAQPVINPMKRLSNIFDQVVDRIEKIESKLAEMQDIPTTSQLVEESYGDKRPAEEMWNN 124
Query 112 VVVEKPFGRDSESSEKLSNDLMEVLNESEIYR 143
+ ++K + +++EK + L ++LN+ + +
Sbjct 125 IKIQKRIEGNEKATEKFTRTLQDLLNDLHVLK 156
> ath:AT1G26540 agenet domain-containing protein
Length=695
Score = 30.8 bits (68), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 23/103 (22%), Positives = 44/103 (42%), Gaps = 15/103 (14%)
Query 15 IAEHLKELSTYFSRQMAPLSSKCDDLVERFRRICTYISGDGYDDAVALKKLSNHLDKLEG 74
+ EH+++ F R + P ++ D+V + DG+ V +KK+
Sbjct 60 LIEHIEQ---RFIRPVPPEENQQKDVVLEEGLLVDADHKDGWWTGVVVKKM--------- 107
Query 75 PEKNNNRLFYLALPPQLFALSVRSIRKH-CWAQKGWNRVVVEK 116
+++N L Y LPP + + +R H W W + +E+
Sbjct 108 --EDDNYLVYFDLPPDIIQFERKQLRTHLIWTGGTWIQPEIEE 148
> xla:414637 stat5b, MGC81286, stat5; signal transducer and activator
of transcription 5B; K11224 signal transducer and activator
of transcription 5B
Length=781
Score = 30.8 bits (68), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 45/97 (46%), Gaps = 9/97 (9%)
Query 86 ALPPQLFALSVRSIRKHCWAQKGWNRVVVEKPFGRDSESSEKLSNDLMEVLNESEIYRID 145
AL Q F + VR H + W+ V E P +D+ + +L L++ L + + +
Sbjct 20 ALYGQHFPIEVRHYLSHWIEAQAWDSVDPENP--QDNLKATQLLEGLVQELQK----KAE 73
Query 146 HYLGKEMTLSIIALRFANVAFKHLFHR---HNVRCVR 179
H +G++ L I L FK+ + R VRC+R
Sbjct 74 HQVGEDGFLLKIKLGHYATQFKNTYERCPMELVRCIR 110
> tgo:TGME49_106620 platelet binding protein GspB, putative
Length=1048
Score = 30.4 bits (67), Expect = 4.2, Method: Composition-based stats.
Identities = 14/45 (31%), Positives = 22/45 (48%), Gaps = 0/45 (0%)
Query 33 LSSKCDDLVERFRRICTYISGDGYDDAVALKKLSNHLDKLEGPEK 77
L S CD V RFRR+ ++ A + + + L + EGP +
Sbjct 682 LLSACDSQVSRFRRVAAFLEEVTCKTGSAAESVCDGLGRAEGPSR 726
> hsa:7038 TG, AITD3, TGN; thyroglobulin; K10809 thyroglobulin
Length=2768
Score = 30.0 bits (66), Expect = 4.4, Method: Composition-based stats.
Identities = 29/91 (31%), Positives = 43/91 (47%), Gaps = 11/91 (12%)
Query 9 EEFWKQIAEHLKELSTYFSRQMAPLSSKCDDLVERFRRICTYISGDGYDDAVALKKLSNH 68
E + Q+AE + S YF+ + P + CDD++E + C I A+ KK+
Sbjct 1900 EHSFCQLAEITESASLYFTCTLYPEAQVCDDIMESNAQGCRLILPQ-MPKALFRKKVI-- 1956
Query 69 LDKLEGPEKNNNRLFYLALPPQ-LFALSVRS 98
LE KN FY LP Q L +S+R+
Sbjct 1957 ---LEDKVKN----FYTRLPFQKLMGISIRN 1980
> dre:445163 zgc:101119
Length=122
Score = 30.0 bits (66), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 18/64 (28%), Positives = 35/64 (54%), Gaps = 5/64 (7%)
Query 125 SEKLSNDLMEVLNESEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRITFKE 184
S+ L+ + +EV+NES ++ + + +++L+F ++ L RH R V T KE
Sbjct 18 SQALNPEHLEVINESHMHAVPPGSESHFKVLVVSLQFEGLS---LLQRH--RLVNETLKE 72
Query 185 DIGT 188
++ T
Sbjct 73 ELST 76
> cel:C18H2.5 hypothetical protein
Length=1139
Score = 29.3 bits (64), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 29/52 (55%), Gaps = 1/52 (1%)
Query 18 HLKELSTYFSRQMAPLSSKCDDLVERFRRICTYISGDGYDDAVALKKLSNHL 69
+L LST + +++ D +VER++RI +GD DD + + KL+ L
Sbjct 83 NLANLSTIITHNPLTVNNHLDGIVERYKRIYESTNGDK-DDLLRILKLNEDL 133
> dre:504079 im:7151855; si:ch211-253b1.6
Length=291
Score = 29.3 bits (64), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 25/102 (24%), Positives = 53/102 (51%), Gaps = 10/102 (9%)
Query 72 LEGPEKNNNRLFYLALPPQLFALSVRSIRKHCWAQKGWNRVVVEKPFGRDSESSEKLSND 131
L+ P+ N + + Y P ++ ++VR + HC+ N ++SES+E L +
Sbjct 167 LDMPDTNVSPVIY---PSEVCVVAVR--QDHCYYSSSSNTDGTNAAGNQESESAEMLRHK 221
Query 132 LMEVLNESEIYR----IDHYLGKEMTLSIIALRFANVAFKHL 169
L VL++ +++R I + + + + +++L+ ANV + L
Sbjct 222 LHVVLSKIKVFRKTMKIKNQMIRRLKSRVLSLK-ANVKTERL 262
Lambda K H
0.322 0.137 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5429324208
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40