bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1336_orf1
Length=185
Score E
Sequences producing significant alignments: (Bits) Value
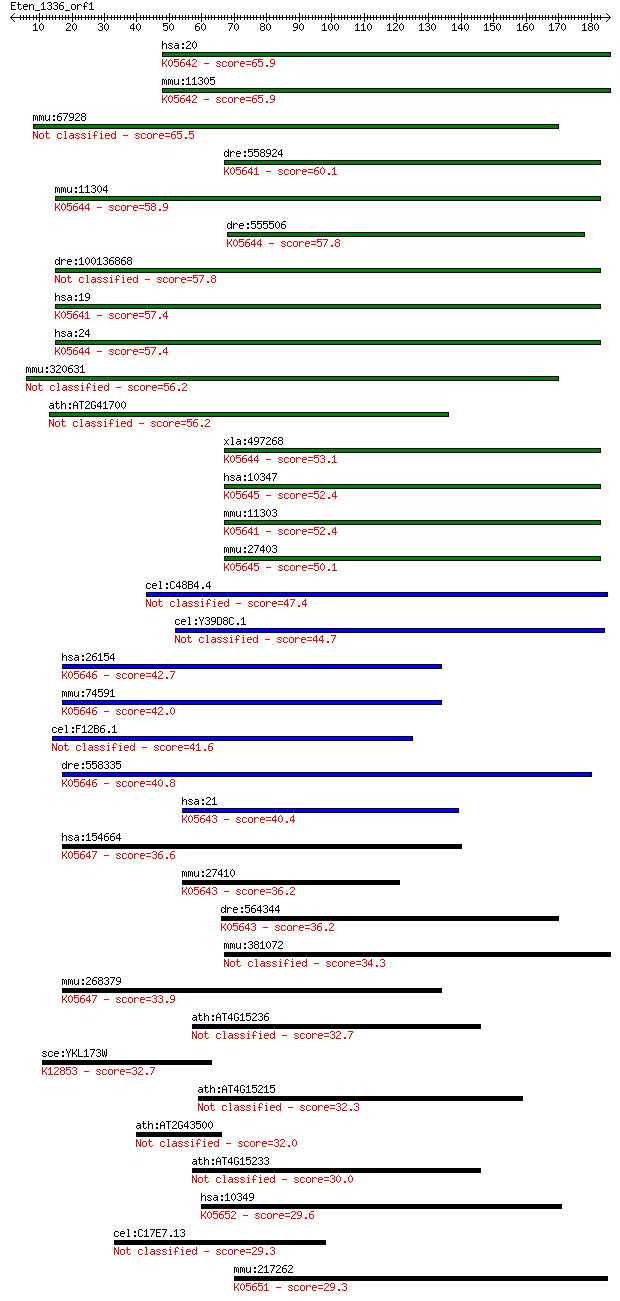
hsa:20 ABCA2, ABC2, MGC129761; ATP-binding cassette, sub-famil... 65.9 7e-11
mmu:11305 Abca2, AI413825, Abc2, D2H0S1474E, mKIAA1062; ATP-bi... 65.9 8e-11
mmu:67928 Abca14, 1700110B15Rik, 4930539G24Rik; ATP-binding ca... 65.5 9e-11
dre:558924 abca1a, abca1, cb939, im:6795541, wu:fj13h01; ATP-b... 60.1 4e-09
mmu:11304 Abca4, AW050280, Abc10, Abcr, D430003I15Rik, RmP; AT... 58.9 1e-08
dre:555506 si:ch211-195i6.4; K05644 ATP-binding cassette, subf... 57.8 2e-08
dre:100136868 abca1b, sb:cb407, wu:fa07e06, zgc:171581; ATP-bi... 57.8 2e-08
hsa:19 ABCA1, ABC-1, ABC1, CERP, FLJ14958, HDLDT1, MGC164864, ... 57.4 2e-08
hsa:24 ABCA4, ABC10, ABCR, ARMD2, CORD3, DKFZp781N1972, FFM, F... 57.4 3e-08
mmu:320631 Abca15, 4930500I12Rik; ATP-binding cassette, sub-fa... 56.2 6e-08
ath:AT2G41700 ATPase, coupled to transmembrane movement of sub... 56.2 7e-08
xla:497268 abca4; ATP-binding cassette, sub-family A (ABC1), m... 53.1 5e-07
hsa:10347 ABCA7, ABCA-SSN, ABCX, FLJ40025; ATP-binding cassett... 52.4 9e-07
mmu:11303 Abca1, Abc1; ATP-binding cassette, sub-family A (ABC... 52.4 9e-07
mmu:27403 Abca7, ABCX, Abc51; ATP-binding cassette, sub-family... 50.1 4e-06
cel:C48B4.4 ced-7; CEll Death abnormality family member (ced-7) 47.4 3e-05
cel:Y39D8C.1 abt-4; ABC Transporter family member (abt-4) 44.7 2e-04
hsa:26154 ABCA12, DKFZp434G232, FLJ41584, ICR2B, LI2; ATP-bind... 42.7 7e-04
mmu:74591 Abca12, 4832428G11Rik, 4833417A11Rik, MGC189862, MGC... 42.0 0.001
cel:F12B6.1 abt-2; ABC Transporter family member (abt-2) 41.6 0.002
dre:558335 abca12, cb352, sb:cb352; ATP-binding cassette, sub-... 40.8 0.002
hsa:21 ABCA3, ABC-C, ABC3, EST111653, LBM180, MGC166979, MGC72... 40.4 0.004
hsa:154664 ABCA13, DKFZp313D2411, FLJ16398, FLJ33876, FLJ33951... 36.6 0.053
mmu:27410 Abca3, 1810036E22Rik, ABC-C, Abc3, MGC90532; ATP-bin... 36.2 0.057
dre:564344 ABC Transporter family member (abt-4)-like; K05643 ... 36.2 0.073
mmu:381072 Abca17; ATP-binding cassette, sub-family A (ABC1), ... 34.3 0.22
mmu:268379 Abca13, 9830132L24, A930002G16Rik, AI956815; ATP-bi... 33.9 0.35
ath:AT4G15236 ATP binding / ATPase/ nucleoside-triphosphatase/... 32.7 0.65
sce:YKL173W SNU114, GIN10; GTPase component of U5 snRNP involv... 32.7 0.77
ath:AT4G15215 PDR13; PDR13; ATP binding / ATPase/ nucleoside-t... 32.3 0.88
ath:AT2G43500 RWP-RK domain-containing protein 32.0 1.3
ath:AT4G15233 ATP binding / ATPase/ nucleoside-triphosphatase/... 30.0 4.4
hsa:10349 ABCA10, EST698739; ATP-binding cassette, sub-family ... 29.6 6.7
cel:C17E7.13 hypothetical protein 29.3 7.7
mmu:217262 Abca9, D630040K07Rik; ATP-binding cassette, sub-fam... 29.3 7.7
> hsa:20 ABCA2, ABC2, MGC129761; ATP-binding cassette, sub-family
A (ABC1), member 2; K05642 ATP-binding cassette, subfamily
A (ABC1), member 2
Length=2436
Score = 65.9 bits (159), Expect = 7e-11, Method: Composition-based stats.
Identities = 36/145 (24%), Positives = 70/145 (48%), Gaps = 7/145 (4%)
Query 48 AVFISNQPFEDND-----DKALDG--IVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQ 100
+ ++N P D L G +V+A F I+A SFV A V+ + E+ K +H Q
Sbjct 1767 GITVTNHPMNKTSASLSLDYLLQGTDVVIAIFIIVAMSFVPASFVVFLVAEKSTKAKHLQ 1826
Query 101 ILARVTPFQYWISSYIVDIILLILPCSIIFGMILWIDVTPLVGPHQRGAFLLGSLLFCLS 160
++ P YW+++Y+ D++ ++P + ++ D+ P A L LL+ S
Sbjct 1827 FVSGCNPIIYWLANYVWDMLNYLVPATCCVIILFVFDLPAYTSPTNFPAVLSLFLLYGWS 1886
Query 161 VCPLGYAISMGVDSPMTFVIVMLLL 185
+ P+ Y S + P + + ++++
Sbjct 1887 ITPIMYPASFWFEVPSSAYVFLIVI 1911
> mmu:11305 Abca2, AI413825, Abc2, D2H0S1474E, mKIAA1062; ATP-binding
cassette, sub-family A (ABC1), member 2; K05642 ATP-binding
cassette, subfamily A (ABC1), member 2
Length=2433
Score = 65.9 bits (159), Expect = 8e-11, Method: Composition-based stats.
Identities = 36/145 (24%), Positives = 70/145 (48%), Gaps = 7/145 (4%)
Query 48 AVFISNQPFEDND-----DKALDG--IVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQ 100
+ ++N P D L G +V+A F I+A SFV A V+ + E+ K +H Q
Sbjct 1766 GITVTNHPMNKTSASLSLDYLLQGTDVVIAIFIIVAMSFVPASFVVFLVAEKSTKAKHLQ 1825
Query 101 ILARVTPFQYWISSYIVDIILLILPCSIIFGMILWIDVTPLVGPHQRGAFLLGSLLFCLS 160
++ P YW+++Y+ D++ ++P + ++ D+ P A L LL+ S
Sbjct 1826 FVSGCNPVIYWLANYVWDMLNYLVPATCCVIILFVFDLPAYTSPTNFPAVLSLFLLYGWS 1885
Query 161 VCPLGYAISMGVDSPMTFVIVMLLL 185
+ P+ Y S + P + + ++++
Sbjct 1886 ITPIMYPASFWFEVPSSAYVFLIVI 1910
> mmu:67928 Abca14, 1700110B15Rik, 4930539G24Rik; ATP-binding
cassette, sub-family A (ABC1), member 14
Length=1683
Score = 65.5 bits (158), Expect = 9e-11, Method: Composition-based stats.
Identities = 41/169 (24%), Positives = 82/169 (48%), Gaps = 7/169 (4%)
Query 8 DATSQLEVTI-WHNSTLRDTLPLHYSHFLNCIAQQRTGVVDAVFISNQP------FEDND 60
D T++ + I W N+ PL S N I + +G + +SN P + ++
Sbjct 1005 DVTTEEKTFIFWFNNEAYHAAPLSLSILDNIIYKYLSGPDATITVSNNPQPQRVTKDKSN 1064
Query 61 DKALDGIVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVDII 120
++++ GI + F + S +G ++ + ERV K +H Q ++ V +W+S+ + D+I
Sbjct 1065 ERSISGIQIVFNLLFGMSIFTSGFCLMTVTERVSKAKHIQFVSGVYTLNFWLSALLWDLI 1124
Query 121 LLILPCSIIFGMILWIDVTPLVGPHQRGAFLLGSLLFCLSVCPLGYAIS 169
+ + C ++ + L+ DV L+ + + +LF S+ P Y +S
Sbjct 1125 IHFVACVLLLVVFLYTDVDILLEKYHFLDTMFILMLFGWSIIPFIYLLS 1173
> dre:558924 abca1a, abca1, cb939, im:6795541, wu:fj13h01; ATP-binding
cassette, sub-family A (ABC1), member 1A; K05641 ATP-binding
cassette, subfamily A (ABC1), member 1
Length=2268
Score = 60.1 bits (144), Expect = 4e-09, Method: Composition-based stats.
Identities = 35/116 (30%), Positives = 58/116 (50%), Gaps = 0/116 (0%)
Query 67 IVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVDIILLILPC 126
++V+ I A SFV A V+ I ERV K +H Q ++ V PF YW+++++ D+ I+P
Sbjct 1662 VLVSICVIFAMSFVPASFVVFLIQERVNKAKHMQFISGVQPFLYWLANFVWDMCNYIVPA 1721
Query 127 SIIFGMILWIDVTPLVGPHQRGAFLLGSLLFCLSVCPLGYAISMGVDSPMTFVIVM 182
+++ + + V L LL+ S+ PL Y S P T +V+
Sbjct 1722 TLVIIIFVCFQQEAYVSSTNLPVLALLLLLYGWSITPLMYPASFFFKIPSTAYVVL 1777
> mmu:11304 Abca4, AW050280, Abc10, Abcr, D430003I15Rik, RmP;
ATP-binding cassette, sub-family A (ABC1), member 4; K05644
ATP-binding cassette, subfamily A (ABC1), member 4
Length=2310
Score = 58.9 bits (141), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 43/176 (24%), Positives = 77/176 (43%), Gaps = 19/176 (10%)
Query 15 VTIWHNSTLRDTLPLHYSHFLNCIAQQRTGVVDAVFISNQPFEDNDDKALD--------G 66
+ + HN+ LR +LP + R + + +QP ++ D
Sbjct 1630 LNVAHNAILRASLP-----------RDRDPEEYGITVISQPLNLTKEQLSDITVLTTSVD 1678
Query 67 IVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVDIILLILPC 126
VVA I A SFV A V+ I ERV K +H Q ++ V+P YW+++++ DI+ +
Sbjct 1679 AVVAICVIFAMSFVPASFVLYLIQERVTKAKHLQFISGVSPTTYWLTNFLWDIMNYAVSA 1738
Query 127 SIIFGMILWIDVTPLVGPHQRGAFLLGSLLFCLSVCPLGYAISMGVDSPMTFVIVM 182
++ G+ + P A + +L+ +V P+ Y S + P T + +
Sbjct 1739 GLVVGIFIGFQKKAYTSPDNLPALVSLLMLYGWAVIPMMYPASFLFEVPSTAYVAL 1794
> dre:555506 si:ch211-195i6.4; K05644 ATP-binding cassette, subfamily
A (ABC1), member 4
Length=2330
Score = 57.8 bits (138), Expect = 2e-08, Method: Composition-based stats.
Identities = 32/110 (29%), Positives = 54/110 (49%), Gaps = 0/110 (0%)
Query 68 VVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVDIILLILPCS 127
VVA I A SF+ A V+ I ERV K +H Q ++ V+P YW +++ D++ + +
Sbjct 1726 VVAICVIFAMSFIPASFVLYLIQERVTKAKHLQFVSGVSPLVYWFANFFWDMLNYSVSTA 1785
Query 128 IIFGMILWIDVTPLVGPHQRGAFLLGSLLFCLSVCPLGYAISMGVDSPMT 177
++ + + D P A + L+ SV P+ Y +S + P T
Sbjct 1786 MVVAIFVGFDKKCYTSPTNLPALIALLCLYGWSVTPMMYPLSYMFNVPST 1835
> dre:100136868 abca1b, sb:cb407, wu:fa07e06, zgc:171581; ATP-binding
cassette, sub-family A (ABC1), member 1B
Length=2271
Score = 57.8 bits (138), Expect = 2e-08, Method: Composition-based stats.
Identities = 46/178 (25%), Positives = 81/178 (45%), Gaps = 23/178 (12%)
Query 15 VTIWHNSTLRDTLP----------LHYSHFLNCIAQQRTGVVDAVFISNQPFEDNDDKAL 64
+ + +N LR LP Y+H LN +Q + V A+ + ++
Sbjct 1615 LNVMNNGILRANLPPGKDPSEFGITAYNHPLNLTKEQLSQV--ALMTT----------SV 1662
Query 65 DGIVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVDIILLIL 124
D ++V+ I A SFV A V+ I ERV K +H Q ++ V P+ YW+++++ D+ ++
Sbjct 1663 D-VLVSICVIFAMSFVPASFVVFLIQERVNKAKHMQFISGVQPYLYWLANFLWDMCNYVV 1721
Query 125 PCSIIFGMILWIDVTPLVGPHQRGAFLLGSLLFCLSVCPLGYAISMGVDSPMTFVIVM 182
P +++ + + V L LL+ S+ PL Y S P T +V+
Sbjct 1722 PATLVILIFVCFQQKAYVSATNLPVLALLLLLYGWSITPLMYPASFLFKIPSTAYVVL 1779
> hsa:19 ABCA1, ABC-1, ABC1, CERP, FLJ14958, HDLDT1, MGC164864,
MGC165011, TGD; ATP-binding cassette, sub-family A (ABC1),
member 1; K05641 ATP-binding cassette, subfamily A (ABC1),
member 1
Length=2261
Score = 57.4 bits (137), Expect = 2e-08, Method: Composition-based stats.
Identities = 49/178 (27%), Positives = 82/178 (46%), Gaps = 23/178 (12%)
Query 15 VTIWHNSTLRDTL-----PLHY-----SHFLNCIAQQRTGVVDAVFISNQPFEDNDDKAL 64
+ + +N+ LR L P HY +H LN QQ + V A+ + ++
Sbjct 1606 LNVINNAILRANLQKGENPSHYGITAFNHPLNLTKQQLSEV--ALMTT----------SV 1653
Query 65 DGIVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVDIILLIL 124
D ++V+ I A SFV A V+ I ERV K +H Q ++ V P YW+S+++ D+ ++
Sbjct 1654 D-VLVSICVIFAMSFVPASFVVFLIQERVSKAKHLQFISGVKPVIYWLSNFVWDMCNYVV 1712
Query 125 PCSIIFGMILWIDVTPLVGPHQRGAFLLGSLLFCLSVCPLGYAISMGVDSPMTFVIVM 182
P +++ + + V L LL+ S+ PL Y S P T +V+
Sbjct 1713 PATLVIIIFICFQQKSYVSSTNLPVLALLLLLYGWSITPLMYPASFVFKIPSTAYVVL 1770
> hsa:24 ABCA4, ABC10, ABCR, ARMD2, CORD3, DKFZp781N1972, FFM,
FLJ17534, RMP, RP19, STGD, STGD1; ATP-binding cassette, sub-family
A (ABC1), member 4; K05644 ATP-binding cassette, subfamily
A (ABC1), member 4
Length=2273
Score = 57.4 bits (137), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 43/176 (24%), Positives = 78/176 (44%), Gaps = 19/176 (10%)
Query 15 VTIWHNSTLRDTLPLHYSHFLNCIAQQRTGVVDAVFISNQPFEDNDDKALD--------G 66
+ + HN+ LR +LP + R+ + + +QP ++ +
Sbjct 1631 LNVAHNAILRASLP-----------KDRSPEEYGITVISQPLNLTKEQLSEITVLTTSVD 1679
Query 67 IVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVDIILLILPC 126
VVA I + SFV A V+ I ERV K +H Q ++ V+P YW+++++ DI+ +
Sbjct 1680 AVVAICVIFSMSFVPASFVLYLIQERVNKSKHLQFISGVSPTTYWVTNFLWDIMNYSVSA 1739
Query 127 SIIFGMILWIDVTPLVGPHQRGAFLLGSLLFCLSVCPLGYAISMGVDSPMTFVIVM 182
++ G+ + P A + LL+ +V P+ Y S D P T + +
Sbjct 1740 GLVVGIFIGFQKKAYTSPENLPALVALLLLYGWAVIPMMYPASFLFDVPSTAYVAL 1795
> mmu:320631 Abca15, 4930500I12Rik; ATP-binding cassette, sub-family
A (ABC1), member 15
Length=1668
Score = 56.2 bits (134), Expect = 6e-08, Method: Composition-based stats.
Identities = 39/168 (23%), Positives = 75/168 (44%), Gaps = 4/168 (2%)
Query 6 ISDATSQLEVTIWHNSTLRDTLPLHYSHFLNCIAQQRTGVVDAVFISNQPFEDNDDK--- 62
I +++ +T+ N+ + L + N + +G ++ + N+P K
Sbjct 1001 IKVVANRVNLTVLFNNEAYHSPSLSLTVLDNILFMSLSGSDASITVFNKPQPSPQRKEWP 1060
Query 63 -ALDGIVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVDIIL 121
+ DG +VAF + + + +G I+ + ER K +H Q L+ V+ YW+S+ + D+I+
Sbjct 1061 GSTDGKIVAFKIQLGMALLVSGFCILTVTERHNKTKHMQFLSGVSILVYWLSALVFDLII 1120
Query 122 LILPCSIIFGMILWIDVTPLVGPHQRGAFLLGSLLFCLSVCPLGYAIS 169
+ C + M + V + +L LF S PL Y +S
Sbjct 1121 FFISCCFLLVMFKYCKFDIYVTDYHILDTMLILTLFGWSAIPLTYLLS 1168
> ath:AT2G41700 ATPase, coupled to transmembrane movement of substances
/ amino acid transmembrane transporter
Length=1846
Score = 56.2 bits (134), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 36/129 (27%), Positives = 61/129 (47%), Gaps = 8/129 (6%)
Query 13 LEVTIWHNSTLRDTLPLHYSHFLNCIAQQRTGVVD-AVFISNQPFEDNDDKA-----LDG 66
L T+ HN T + P++ + I + TG + + N P + LD
Sbjct 1038 LGYTVLHNGTCQHAGPIYINVMHAAILRLATGNKNMTIQTRNHPLPPTKTQRIQRHDLDA 1097
Query 67 IVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVDIILLILPC 126
A IA+SF+ A + + ER K +HQQ+++ V+ YW+S+Y+ D I + P
Sbjct 1098 FSAAIIVNIAFSFIPASFAVPIVKEREVKAKHQQLISGVSVLSYWLSTYVWDFISFLFPS 1157
Query 127 SIIFGMILW 135
+ F +IL+
Sbjct 1158 T--FAIILF 1164
> xla:497268 abca4; ATP-binding cassette, sub-family A (ABC1),
member 4; K05644 ATP-binding cassette, subfamily A (ABC1),
member 4
Length=2363
Score = 53.1 bits (126), Expect = 5e-07, Method: Composition-based stats.
Identities = 31/116 (26%), Positives = 53/116 (45%), Gaps = 0/116 (0%)
Query 67 IVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVDIILLILPC 126
+VVA I A SF+ A V+ I ERV +H Q ++ VTP YW++++ DI+ L
Sbjct 1759 VVVAICVIFAMSFIPASFVLYLIQERVSNAKHLQFVSGVTPAIYWLTNFAWDIVNYALSV 1818
Query 127 SIIFGMILWIDVTPLVGPHQRGAFLLGSLLFCLSVCPLGYAISMGVDSPMTFVIVM 182
+++ + D A + + +V P+ Y S + P T + +
Sbjct 1819 AMVVIIFFAFDKKAYTSSTNLPALIALLFFYGWAVIPMMYPASYLFNVPSTAYVAL 1874
> hsa:10347 ABCA7, ABCA-SSN, ABCX, FLJ40025; ATP-binding cassette,
sub-family A (ABC1), member 7; K05645 ATP-binding cassette,
subfamily A (ABC1), member 7
Length=2146
Score = 52.4 bits (124), Expect = 9e-07, Method: Composition-based stats.
Identities = 35/116 (30%), Positives = 59/116 (50%), Gaps = 0/116 (0%)
Query 67 IVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVDIILLILPC 126
++V+ + A SFV A +V I ERV + +H Q++ ++P YW+ +++ D+ ++P
Sbjct 1536 VLVSICVVFAMSFVPASFTLVLIEERVTRAKHLQLMGGLSPTLYWLGNFLWDMCNYLVPA 1595
Query 127 SIIFGMILWIDVTPLVGPHQRGAFLLGSLLFCLSVCPLGYAISMGVDSPMTFVIVM 182
I+ + L V P A LL LL+ S+ PL Y S P T +V+
Sbjct 1596 CIVVLIFLAFQQRAYVAPANLPALLLLLLLYGWSITPLMYPASFFFSVPSTAYVVL 1651
> mmu:11303 Abca1, Abc1; ATP-binding cassette, sub-family A (ABC1),
member 1; K05641 ATP-binding cassette, subfamily A (ABC1),
member 1
Length=2261
Score = 52.4 bits (124), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 34/116 (29%), Positives = 57/116 (49%), Gaps = 0/116 (0%)
Query 67 IVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVDIILLILPC 126
++V+ I A SFV A V+ I ERV K +H Q ++ V P YW+S+++ D+ ++P
Sbjct 1655 VLVSICVIFAMSFVPASFVVFLIQERVSKAKHLQFISGVKPVIYWLSNFVWDMCNYVVPA 1714
Query 127 SIIFGMILWIDVTPLVGPHQRGAFLLGSLLFCLSVCPLGYAISMGVDSPMTFVIVM 182
+++ + + V L LL+ S+ PL Y S P T +V+
Sbjct 1715 TLVIIIFICFQQKSYVSSTNLPVLALLLLLYGWSITPLMYPASFVFKIPSTAYVVL 1770
> mmu:27403 Abca7, ABCX, Abc51; ATP-binding cassette, sub-family
A (ABC1), member 7; K05645 ATP-binding cassette, subfamily
A (ABC1), member 7
Length=2159
Score = 50.1 bits (118), Expect = 4e-06, Method: Composition-based stats.
Identities = 32/116 (27%), Positives = 57/116 (49%), Gaps = 0/116 (0%)
Query 67 IVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVDIILLILPC 126
++V+ + A SFV A +V I ER+ + +H Q+++ + YW+ +++ D+ ++
Sbjct 1550 VLVSICVVFAMSFVPASFTLVLIEERITRAKHLQLVSGLPQTLYWLGNFLWDMCNYLVAV 1609
Query 127 SIIFGMILWIDVTPLVGPHQRGAFLLGSLLFCLSVCPLGYAISMGVDSPMTFVIVM 182
I+ + L V P A LL LL+ S+ PL Y S P T +V+
Sbjct 1610 CIVVFIFLAFQQRAYVAPENLPALLLLLLLYGWSITPLMYPASFFFSVPSTAYVVL 1665
> cel:C48B4.4 ced-7; CEll Death abnormality family member (ced-7)
Length=1689
Score = 47.4 bits (111), Expect = 3e-05, Method: Composition-based stats.
Identities = 42/153 (27%), Positives = 72/153 (47%), Gaps = 19/153 (12%)
Query 43 TGVVDA-----VFISNQPFEDND--DKALDGIVVAFFTIIAYSFVAAGVVIVCIVERVRK 95
TG VDA VF+ ++ +++ L +++A I+ ++ V + V+ I ER +
Sbjct 1078 TGTVDAEISSGVFLYSKSTSNSNLLPSQLIDVLLAPMLILIFAMVTSTFVMFLIEERTCQ 1137
Query 96 VRHQQILARVTPFQYWISSYIVDIILLILPCSIIFGMIL---WI-DVTPLVGPHQRGAFL 151
HQQ L ++P ++ +S I D IL L C I M L W+ D +V +
Sbjct 1138 FAHQQFLTGISPITFYSASLIYDGILYSLICLIFLFMFLAFHWMYDHLAIV--------I 1189
Query 152 LGSLLFCLSVCPLGYAISMGVDSPMTFVIVMLL 184
L L+ S P YA+S SP +++++
Sbjct 1190 LFWFLYFFSSVPFIYAVSFLFQSPSKANVLLII 1222
> cel:Y39D8C.1 abt-4; ABC Transporter family member (abt-4)
Length=1802
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 36/139 (25%), Positives = 69/139 (49%), Gaps = 11/139 (7%)
Query 52 SNQPFEDNDDKALDG--IVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQ 109
S Q N +++ DG ++A+ I++++ AG I ER +K +H Q+L+ + P+
Sbjct 1141 STQDTLKNTNRS-DGAAFLIAYGLIVSFAVCVAGYSQFLITERKKKSKHMQLLSGIRPWM 1199
Query 110 YWISSYIVDIILLILPCSIIFGMILWI-DVTPLVGPHQRGAFL---LGSLLFCLSVCPLG 165
+W++++I D ++ + F I +I ++T H G L L LL+ + P
Sbjct 1200 FWLTAFIWDAAWFVIRI-LCFDAIFYIFNITAYT--HDFGVMLILTLSFLLYGWTALPFT 1256
Query 166 YAISMGVDS-PMTFVIVML 183
Y +S P F++V +
Sbjct 1257 YWFQFFFESAPKGFMMVTM 1275
> hsa:26154 ABCA12, DKFZp434G232, FLJ41584, ICR2B, LI2; ATP-binding
cassette, sub-family A (ABC1), member 12; K05646 ATP-binding
cassette, subfamily A (ABC1), member 12
Length=2277
Score = 42.7 bits (99), Expect = 7e-04, Method: Composition-based stats.
Identities = 26/127 (20%), Positives = 58/127 (45%), Gaps = 10/127 (7%)
Query 17 IWHNSTLRDTLPLHYSHFLNCIAQQRTGVVDA----VFISNQPFEDNDDK------ALDG 66
+W++ +LP + + N + + DA + + + P+ D+ +L
Sbjct 1609 VWYDPEGYHSLPAYLNSLNNFLLRVNMSKYDAARHGIIMYSHPYPGVQDQEQATISSLID 1668
Query 67 IVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVDIILLILPC 126
I+VA ++ YS A V + E K + Q ++ + YW++++I D++ ++P
Sbjct 1669 ILVALSILMGYSVTTASFVTYVVREHQTKAKQLQHISGIGVTCYWVTNFIYDMVFYLVPV 1728
Query 127 SIIFGMI 133
+ G+I
Sbjct 1729 AFSIGII 1735
> mmu:74591 Abca12, 4832428G11Rik, 4833417A11Rik, MGC189862, MGC189975;
ATP-binding cassette, sub-family A (ABC1), member
12; K05646 ATP-binding cassette, subfamily A (ABC1), member
12
Length=2595
Score = 42.0 bits (97), Expect = 0.001, Method: Composition-based stats.
Identities = 26/127 (20%), Positives = 58/127 (45%), Gaps = 10/127 (7%)
Query 17 IWHNSTLRDTLPLHYSHFLNCIAQQRTGVVDA----VFISNQPFEDNDDK------ALDG 66
+W++ +LP + + N + + DA + + + P+ D+ +L
Sbjct 1927 VWYDPEGYHSLPAYLNSLNNFLLRVNMSEYDAARHGIIMYSHPYPGVQDQEQATISSLID 1986
Query 67 IVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVDIILLILPC 126
I+VA ++ YS A V + E K + Q ++ + YW++++I D++ ++P
Sbjct 1987 ILVALSILMGYSVTTASFVTYIVREHQTKAKQLQHISGIGVTCYWVTNFIYDMVFYLVPV 2046
Query 127 SIIFGMI 133
+ G+I
Sbjct 2047 AFSIGVI 2053
> cel:F12B6.1 abt-2; ABC Transporter family member (abt-2)
Length=2146
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/125 (20%), Positives = 52/125 (41%), Gaps = 14/125 (11%)
Query 14 EVTIWHNSTLRDTLPLHYSHFLNCIAQQRTGVVD----AVFISNQPFEDNDDKALDGIVV 69
V +W N+ + P+ + N + +Q +D + N P + LD +
Sbjct 1455 NVKVWFNNKIWPGFPIASNILSNALLRQEDYAIDPEDLGILTMNHPMNKTISQTLDQNAL 1514
Query 70 AFFTIIA----------YSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVDI 119
F +A S + AG + + +R+ + H Q++ + YW++SY+ D+
Sbjct 1515 KFTQALAVFRITILLLVLSMIPAGFTVYLVEDRICEALHLQLVGGLRKVTYWVTSYLYDM 1574
Query 120 ILLIL 124
++ L
Sbjct 1575 VVYTL 1579
> dre:558335 abca12, cb352, sb:cb352; ATP-binding cassette, sub-family
A (ABC1), member 12; K05646 ATP-binding cassette, subfamily
A (ABC1), member 12
Length=3644
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 46/175 (26%), Positives = 74/175 (42%), Gaps = 13/175 (7%)
Query 17 IWHNSTLRDTLPLHYSH----FLNCIAQQRTGVVDAVFISNQPFE---DNDDKALDGIV- 68
+W+NS ++P + + FL A+ IS+ P+ ++D + G+V
Sbjct 2977 VWYNSEGHHSMPAYLNSLNNFFLRSSLPPEKRYQYAISISSHPYPGQVQDEDLMVGGLVS 3036
Query 69 --VAFFTIIAYSFVAAGVVIVCIVER-VRKVRHQQILARVTPFQYWISSYIVDIILLILP 125
VA + YS + A VI + E R QQI PF YWI ++ D+ L ++P
Sbjct 3037 ILVALCVLTGYSIMTASFVIYEVQEHHTGSKRLQQISGISEPF-YWIINFFYDMALYMIP 3095
Query 126 CSIIFGMILWIDVTPLVGPHQRGAFLLGSLLFCLSVCPLGYAIS-MGVDSPMTFV 179
+ M+ + A L +LF S P Y +S M D+ M F+
Sbjct 3096 VVLSVVMVAAFQLPAFTERQNLAAVTLLLVLFGFSTFPWMYLLSAMFKDTEMAFI 3150
> hsa:21 ABCA3, ABC-C, ABC3, EST111653, LBM180, MGC166979, MGC72201,
SMDP3; ATP-binding cassette, sub-family A (ABC1), member
3; K05643 ATP-binding cassette, subfamily A (ABC1), member
3
Length=1704
Score = 40.4 bits (93), Expect = 0.004, Method: Composition-based stats.
Identities = 20/85 (23%), Positives = 44/85 (51%), Gaps = 0/85 (0%)
Query 54 QPFEDNDDKALDGIVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWIS 113
Q +D ++ G +A + A +F+A+ I+ + ER + +H Q ++ V +W+S
Sbjct 1085 QAAKDQFNEGRKGFDIALNLLFAMAFLASTFSILAVSERAVQAKHVQFVSGVHVASFWLS 1144
Query 114 SYIVDIILLILPCSIIFGMILWIDV 138
+ + D+I ++P ++ + DV
Sbjct 1145 ALLWDLISFLIPSLLLLVVFKAFDV 1169
> hsa:154664 ABCA13, DKFZp313D2411, FLJ16398, FLJ33876, FLJ33951;
ATP-binding cassette, sub-family A (ABC1), member 13; K05647
ATP binding cassette, subfamily A (ABC1), member 13
Length=5058
Score = 36.6 bits (83), Expect = 0.053, Method: Composition-based stats.
Identities = 28/134 (20%), Positives = 62/134 (46%), Gaps = 11/134 (8%)
Query 17 IWHNSTLRDTLPLHYSHFLNCIA-QQRTGVVD----AVFISNQPFED---NDDKALDGIV 68
+W+N +LP + +H N I Q VD + + + P+ N DK L+ I
Sbjct 4395 VWYNQKGFHSLPSYLNHLNNLILWQHLPPTVDWRQYGITLYSHPYGGALLNKDKILESIR 4454
Query 69 ---VAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVDIILLILP 125
VA ++ +S ++A + + +RV + Q ++ + YW ++++ D++ ++
Sbjct 4455 QCGVALCIVLGFSILSASIGSSVVRDRVIGAKRLQHISGLGYRMYWFTNFLYDMLFYLVS 4514
Query 126 CSIIFGMILWIDVT 139
+ +I+ +T
Sbjct 4515 VCLCVAVIVAFQLT 4528
> mmu:27410 Abca3, 1810036E22Rik, ABC-C, Abc3, MGC90532; ATP-binding
cassette, sub-family A (ABC1), member 3; K05643 ATP-binding
cassette, subfamily A (ABC1), member 3
Length=1704
Score = 36.2 bits (82), Expect = 0.057, Method: Composition-based stats.
Identities = 18/67 (26%), Positives = 37/67 (55%), Gaps = 0/67 (0%)
Query 54 QPFEDNDDKALDGIVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWIS 113
Q +D+ ++ G +A +IA +F+A+ I+ + ER + +H Q ++ V +W S
Sbjct 1085 QVAKDHFNEGRKGFDIALNLLIAMAFLASTFSILAVSERAVQAKHVQFVSGVHVATFWFS 1144
Query 114 SYIVDII 120
+ + D+I
Sbjct 1145 ALLWDLI 1151
> dre:564344 ABC Transporter family member (abt-4)-like; K05643
ATP-binding cassette, subfamily A (ABC1), member 3
Length=1343
Score = 36.2 bits (82), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 26/104 (25%), Positives = 49/104 (47%), Gaps = 0/104 (0%)
Query 66 GIVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVDIILLILP 125
G +A + + +A+ ++ + ER K +H Q ++ V +W S+ + D+I +LP
Sbjct 734 GFAIAINLMYGMASLASTFALLLVSERSVKSKHVQQVSGVYLTNFWFSALLWDLINFLLP 793
Query 126 CSIIFGMILWIDVTPLVGPHQRGAFLLGSLLFCLSVCPLGYAIS 169
C ++ + V V + L LL+ +V PL Y +S
Sbjct 794 CLLMLVVFRVFSVKAFVAQNHLVDVLFLLLLYGWAVIPLMYLMS 837
> mmu:381072 Abca17; ATP-binding cassette, sub-family A (ABC1),
member 17
Length=1733
Score = 34.3 bits (77), Expect = 0.22, Method: Composition-based stats.
Identities = 27/119 (22%), Positives = 61/119 (51%), Gaps = 2/119 (1%)
Query 67 IVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVDIILLILPC 126
+VV F IA F+++ I+ + E+ K + Q ++ V+ +W+S+ + D+I ++P
Sbjct 1082 LVVNFLFGIA--FLSSSFSILTVGEKSVKSKSLQFVSGVSTAVFWLSALLWDLISFLVPT 1139
Query 127 SIIFGMILWIDVTPLVGPHQRGAFLLGSLLFCLSVCPLGYAISMGVDSPMTFVIVMLLL 185
++ + LW A +L +L+ +V PL Y +S ++P + + ++++
Sbjct 1140 LLLVLVFLWYKEEAFAHHESIPAVVLIMMLYGWAVIPLVYTVSFSFNTPGSACVKLVVM 1198
> mmu:268379 Abca13, 9830132L24, A930002G16Rik, AI956815; ATP-binding
cassette, sub-family A (ABC1), member 13; K05647 ATP
binding cassette, subfamily A (ABC1), member 13
Length=5034
Score = 33.9 bits (76), Expect = 0.35, Method: Composition-based stats.
Identities = 25/129 (19%), Positives = 59/129 (45%), Gaps = 12/129 (9%)
Query 17 IWHNSTLRDTLPLHYSHFLNCIAQQR--TGVVD----AVFISNQPFED---NDDKALDGI 67
+W+N +LP + +H N I VD + + + P+ N+D+ L+ I
Sbjct 4368 VWYNQKGFHSLPSYLNHLNNLILWHHLPANAVDWRQYGITLYSHPYGGALLNEDRILESI 4427
Query 68 V---VAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVDIILLIL 124
VA ++ +S ++A + + +RV + Q ++ + YW+ +++ D++ ++
Sbjct 4428 RQCGVALCIVLGFSILSASIGSSVVRDRVTGAKRLQHISGLGHRTYWLINFLYDMLFYLV 4487
Query 125 PCSIIFGMI 133
+ +I
Sbjct 4488 SVCLCVAVI 4496
> ath:AT4G15236 ATP binding / ATPase/ nucleoside-triphosphatase/
nucleotide binding
Length=1388
Score = 32.7 bits (73), Expect = 0.65, Method: Composition-based stats.
Identities = 23/90 (25%), Positives = 46/90 (51%), Gaps = 6/90 (6%)
Query 57 EDNDDKALDGIVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARV-TPFQYWISSY 115
E N+ + L + + FT++ +S + ++ V R V +++ +R+ P+ Y ++
Sbjct 1157 EINNQQDLFNVFGSMFTVVLFSGINNCSTVIFCVATERNVFYRERFSRMYNPWAYSLAQV 1216
Query 116 IVDIILLILPCSIIFGMILWIDVTPLVGPH 145
+V+I P S+ +I I V P+VG H
Sbjct 1217 LVEI-----PYSLFQSIIYVIIVYPMVGYH 1241
> sce:YKL173W SNU114, GIN10; GTPase component of U5 snRNP involved
in mRNA splicing via spliceosome; binds directly to U5
snRNA; proposed to be involved in conformational changes of
the spliceosome; similarity to ribosomal translocation factor
EF-2; K12853 114 kDa U5 small nuclear ribonucleoprotein component
Length=1008
Score = 32.7 bits (73), Expect = 0.77, Method: Composition-based stats.
Identities = 19/52 (36%), Positives = 26/52 (50%), Gaps = 1/52 (1%)
Query 11 SQLEVTIWHNSTLRDTLPLHYSHFLNCIAQQRTGVVDAVFISNQPFEDNDDK 62
S V + + D P H L I +Q+TG+VDA+ QPFE D+K
Sbjct 402 SNFRVNLSQEALQYDPQPF-LKHVLQLIFRQQTGLVDAITRCYQPFELFDNK 452
> ath:AT4G15215 PDR13; PDR13; ATP binding / ATPase/ nucleoside-triphosphatase/
nucleotide binding
Length=1390
Score = 32.3 bits (72), Expect = 0.88, Method: Composition-based stats.
Identities = 24/102 (23%), Positives = 51/102 (50%), Gaps = 6/102 (5%)
Query 59 NDDKALDGIVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVD 118
N+ + L I + +TI+ +S + ++ + R V +++ AR+ + W +Y
Sbjct 1161 NNQQDLFSIFGSMYTIVIFSGINNCATVMNFIATERNVFYRERFARM--YSSW--AYSFS 1216
Query 119 IILLILPCSIIFGMILWIDVTPLVGPHQRGAFLLGSL--LFC 158
+L+ +P S++ ++ I V P++G H + SL +FC
Sbjct 1217 QVLVEVPYSLLQSLLCTIIVYPMIGYHMSVYKMFWSLYSIFC 1258
> ath:AT2G43500 RWP-RK domain-containing protein
Length=947
Score = 32.0 bits (71), Expect = 1.3, Method: Composition-based stats.
Identities = 13/26 (50%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 40 QQRTGVVDAVFISNQPFEDNDDKALD 65
+++ G+V FISNQPF +D KA D
Sbjct 409 REKEGIVGKAFISNQPFFSSDVKAYD 434
> ath:AT4G15233 ATP binding / ATPase/ nucleoside-triphosphatase/
nucleotide binding
Length=1311
Score = 30.0 bits (66), Expect = 4.4, Method: Composition-based stats.
Identities = 21/89 (23%), Positives = 46/89 (51%), Gaps = 4/89 (4%)
Query 57 EDNDDKALDGIVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYI 116
E N+ + L + + FT++ +S + ++ V R V +++ +R+ + W +Y
Sbjct 1080 EINNQQDLFNVFGSMFTVVLFSGINNCSTVLFSVATERNVFYRERFSRM--YNSW--AYS 1135
Query 117 VDIILLILPCSIIFGMILWIDVTPLVGPH 145
+ +L+ +P S+ ++ I V P+VG H
Sbjct 1136 LAQVLVEIPYSLFQSIVYVIIVYPMVGYH 1164
> hsa:10349 ABCA10, EST698739; ATP-binding cassette, sub-family
A (ABC1), member 10; K05652 ATP-binding cassette, subfamily
A (ABC1), member 10
Length=1543
Score = 29.6 bits (65), Expect = 6.7, Method: Composition-based stats.
Identities = 32/113 (28%), Positives = 52/113 (46%), Gaps = 11/113 (9%)
Query 60 DDKALD--GIVVAFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIV 117
DD LD I + F ++ + V+ + + I + + V+ Q ++ + P YW +V
Sbjct 924 DDIVLDLGFIDGSIFLLLITNCVSPFIGMSSISDYKKNVQSQLWISGLWPSAYWCGQALV 983
Query 118 DIILLILPCSIIFGMILWIDVTPLVGPHQRGAFLLGSLLFCLSVCPLGYAISM 170
DI L L I+F + L L G L L+F L VC +G A+S+
Sbjct 984 DIPLYFL---ILFSIHLIYYFIFL------GFQLSWELMFVLVVCIIGCAVSL 1027
> cel:C17E7.13 hypothetical protein
Length=284
Score = 29.3 bits (64), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 33/67 (49%), Gaps = 10/67 (14%)
Query 33 HFLNCIAQQRTGVVDAVFISNQPFEDNDDKALDGIVVAFFTIIAYSFVAAGVVIV--CIV 90
H L + +Q VV+ FI D A+ V + ++ Y F+AA +++V C+V
Sbjct 143 HVLYVLEKQFGHVVEMKFIC--------DPAIVKDQVENYVLLIYGFIAAALILVSLCVV 194
Query 91 ERVRKVR 97
R RK R
Sbjct 195 SRARKRR 201
> mmu:217262 Abca9, D630040K07Rik; ATP-binding cassette, sub-family
A (ABC1), member 9; K05651 ATP-binding cassette, subfamily
A (ABC1), member 9
Length=1623
Score = 29.3 bits (64), Expect = 7.7, Method: Composition-based stats.
Identities = 29/115 (25%), Positives = 50/115 (43%), Gaps = 11/115 (9%)
Query 70 AFFTIIAYSFVAAGVVIVCIVERVRKVRHQQILARVTPFQYWISSYIVDIILLILPCSII 129
AFF I + + + + I + +KV Q + + P YW +VDI I
Sbjct 1025 AFFWIPVAASLTPYIAMGSISDHKKKVLSQLWTSGLYPSAYWCGQALVDI-------PIY 1077
Query 130 FGMILWIDVTPLVGPHQRGAFLLGSLLFCLSVCPLGYAISMGVDSPMTFVIVMLL 184
F ++ + + V + ++ SLL + C +GYA S+ MT+VI +
Sbjct 1078 FLILFLMQIMDSVFSSEEFISVMESLLIQIP-CSIGYASSLIF---MTYVISFIF 1128
Lambda K H
0.330 0.144 0.451
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5106150148
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40