bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1312_orf1
Length=183
Score E
Sequences producing significant alignments: (Bits) Value
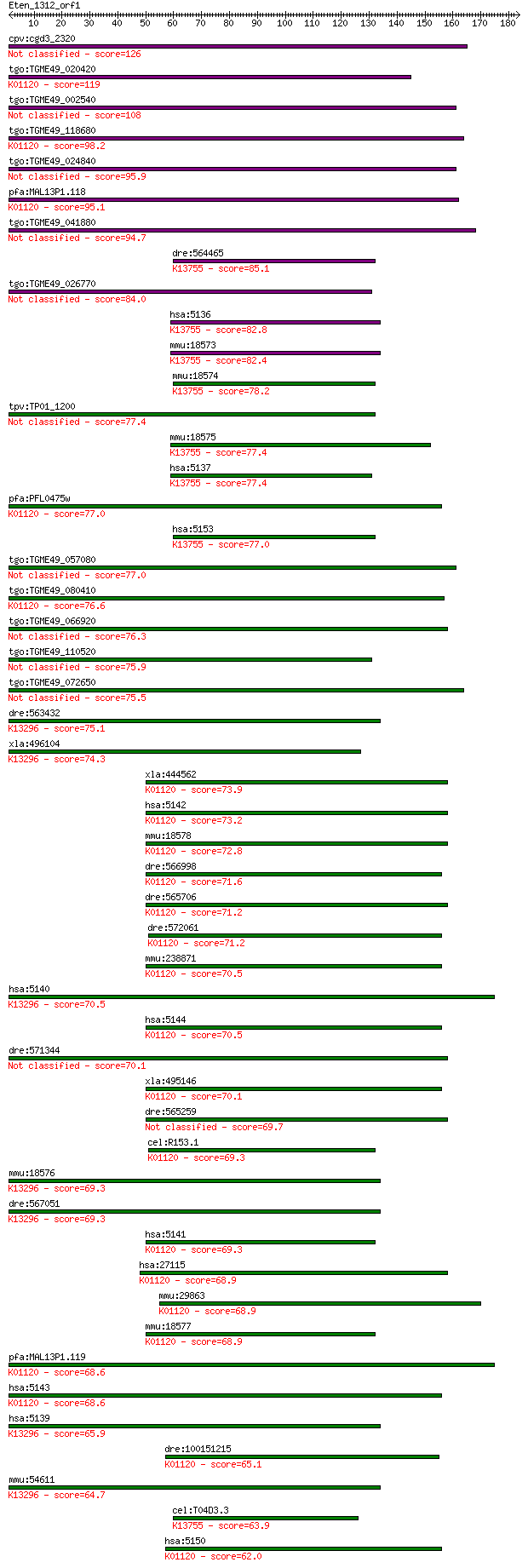
cpv:cgd3_2320 cGMP phosphodiesterase A4 126 3e-29
tgo:TGME49_020420 3'5'-cyclic nucleotide phosphodiesterase, pu... 119 4e-27
tgo:TGME49_002540 3',5'-cyclic nucleotide phosphodiesterase, p... 108 1e-23
tgo:TGME49_118680 3',5'--cyclic-nucleotide phosphodiesterase, ... 98.2 1e-20
tgo:TGME49_024840 cAMP phosphodiesterase, putative (EC:3.1.4.1... 95.9 7e-20
pfa:MAL13P1.118 3',5'-cyclic nucleotide phosphodiesterase (EC:... 95.1 1e-19
tgo:TGME49_041880 3', 5'-cyclic nucleotide phosphodiesterase, ... 94.7 2e-19
dre:564465 pde1a; phosphodiesterase 1A, calmodulin-dependent; ... 85.1 1e-16
tgo:TGME49_026770 cGMP-inhibited 3',5'-cyclic phosphodiesteras... 84.0 2e-16
hsa:5136 PDE1A, HCAM1, HSPDE1A, MGC26303; phosphodiesterase 1A... 82.8 7e-16
mmu:18573 Pde1a, AI987702, AW125737, MGC116577; phosphodiester... 82.4 8e-16
mmu:18574 Pde1b, 63kDa, Pde1b1; phosphodiesterase 1B, Ca2+-cal... 78.2 2e-14
tpv:TP01_1200 hypothetical protein 77.4 2e-14
mmu:18575 Pde1c; phosphodiesterase 1C (EC:3.1.4.17); K13755 ca... 77.4 2e-14
hsa:5137 PDE1C, Hcam3; phosphodiesterase 1C, calmodulin-depend... 77.4 3e-14
pfa:PFL0475w PDE1; cGMP-specific phosphodiesterase (EC:3.1.4.1... 77.0 3e-14
hsa:5153 PDE1B, PDE1B1, PDES1B; phosphodiesterase 1B, calmodul... 77.0 3e-14
tgo:TGME49_057080 cAMP-specific 3',5'-cyclic phosphodiesterase... 77.0 4e-14
tgo:TGME49_080410 3',5'-cyclic phosphodiesterase, putative (EC... 76.6 5e-14
tgo:TGME49_066920 phosphodiesterase, putative (EC:3.1.4.17) 76.3 6e-14
tgo:TGME49_110520 calcium/calmodulin-dependent 3', 5'-cyclic n... 75.9 8e-14
tgo:TGME49_072650 calcium/calmodulin-dependent 3',5'-cyclic nu... 75.5 1e-13
dre:563432 pde3b; phosphodiesterase 3B, cGMP-inhibited; K13296... 75.1 1e-13
xla:496104 pde3b; phosphodiesterase 3b, cGMP-inhibited; K13296... 74.3 2e-13
xla:444562 pde4b, MGC83972; phosphodiesterase 4B, cAMP-specifi... 73.9 3e-13
hsa:5142 PDE4B, DKFZp686F2182, DPDE4, MGC126529, PDE4B5, PDEIV... 73.2 5e-13
mmu:18578 Pde4b, Dpde4, R74983, dunce; phosphodiesterase 4B, c... 72.8 6e-13
dre:566998 novel protein similar to vertebrate phosphodiestera... 71.6 1e-12
dre:565706 cAMP-specific 3,5-cyclic phosphodiesterase 4B-like;... 71.2 2e-12
dre:572061 pde4a; phosphodiesterase 4A, cAMP-specific; K01120 ... 71.2 2e-12
mmu:238871 Pde4d, 9630011N22Rik, Dpde3; phosphodiesterase 4D, ... 70.5 3e-12
hsa:5140 PDE3B, HcGIP1, cGIPDE1; phosphodiesterase 3B, cGMP-in... 70.5 3e-12
hsa:5144 PDE4D, DKFZp686M11213, DPDE3, FLJ97311, HSPDE4D, PDE4... 70.5 3e-12
dre:571344 fc68c05, si:ch211-255d18.8, wu:fc68c05; si:ch211-25... 70.1 4e-12
xla:495146 hypothetical LOC495146; K01120 3',5'-cyclic-nucleot... 70.1 4e-12
dre:565259 pde4c, im:7160317, si:dkey-149i17.5; phosphodiester... 69.7 5e-12
cel:R153.1 pde-4; PhosphoDiEsterase family member (pde-4); K01... 69.3 6e-12
mmu:18576 Pde3b, 9830102A01Rik, AI847709; phosphodiesterase 3B... 69.3 6e-12
dre:567051 si:dkey-48h7.2; K13296 cGMP-inhibited 3',5'-cyclic ... 69.3 7e-12
hsa:5141 PDE4A, DPDE2, PDE4, PDE46; phosphodiesterase 4A, cAMP... 69.3 7e-12
hsa:27115 PDE7B, MGC88256, bA472E5.1; phosphodiesterase 7B (EC... 68.9 9e-12
mmu:29863 Pde7b; phosphodiesterase 7B (EC:3.1.4.17); K01120 3'... 68.9 1e-11
mmu:18577 Pde4a, D9Ertd60e, Dpde2; phosphodiesterase 4A, cAMP ... 68.9 1e-11
pfa:MAL13P1.119 calcium/calmodulin-dependent 3',5'-cyclic nucl... 68.6 1e-11
hsa:5143 PDE4C, DPDE1, MGC126222; phosphodiesterase 4C, cAMP-s... 68.6 1e-11
hsa:5139 PDE3A, CGI-PDE; phosphodiesterase 3A, cGMP-inhibited ... 65.9 7e-11
dre:100151215 phosphodiesterase 7A-like (EC:3.1.4.17); K01120 ... 65.1 1e-10
mmu:54611 Pde3a, A930022O17Rik, C87899; phosphodiesterase 3A, ... 64.7 2e-10
cel:T04D3.3 pde-1; PhosphoDiEsterase family member (pde-1); K1... 63.9 3e-10
hsa:5150 PDE7A, HCP1, PDE7; phosphodiesterase 7A (EC:3.1.4.17)... 62.0 1e-09
> cpv:cgd3_2320 cGMP phosphodiesterase A4
Length=997
Score = 126 bits (317), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 59/164 (35%), Positives = 96/164 (58%), Gaps = 23/164 (14%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
D+ HF+ +++FR+ ++ +D+ +P+ D D LA+ C+
Sbjct 840 DMHRHFECVSRFRV------------RRQAVDW------DPYG----DAQDRLMLARTCL 877
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFLK 120
+ AD+GH A+KW QHYKW ++++ EFF QG +E L L +SP+CD +TDVPKSQ+GFL
Sbjct 878 KAADIGHGALKWNQHYKWCRSVVEEFFLQGDEEKALSLPISPICDRESTDVPKSQVGFLN 937
Query 121 LICFPLFESLAQADNSGDLAAICLSFLKDNSETWQRISDSEMDW 164
+C PLF+ L D GD+ C+ + +N W+ I+++ + W
Sbjct 938 FVCLPLFQELCYVDVEGDVRR-CIDRILENINNWEDIAEAGIQW 980
> tgo:TGME49_020420 3'5'-cyclic nucleotide phosphodiesterase,
putative (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=466
Score = 119 bits (299), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 67/155 (43%), Positives = 94/155 (60%), Gaps = 11/155 (7%)
Query 1 DLSTHFDFLAQFRLQLHFLNS-KVNSEKKSEIDF-KSPQTL--NPHKTL-------KTDP 49
D+ HFDF+ +FRL+ K+ E+ + F ++P +L +P +T K +
Sbjct 308 DILFHFDFIGRFRLRRPMSPFFKLLGERGANPSFGQNPVSLLSSPSRTATDVEATSKEEA 367
Query 50 ADCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAAT 109
D LAKACIR ADVGH+AV+W QHY +S+A+ EFF QG+ E L L++SP CD +T
Sbjct 368 QDLTLLAKACIRSADVGHAAVEWRQHYFYSKAVQTEFFNQGKAEKRLRLRISPCCDPVST 427
Query 110 DVPKSQIGFLKLICFPLFESLAQADNSGDLAAICL 144
DVPK Q GF+ I PLFE LA + +G + +CL
Sbjct 428 DVPKCQDGFISYISRPLFEELALINTNGAIMQVCL 462
> tgo:TGME49_002540 3',5'-cyclic nucleotide phosphodiesterase,
putative (EC:3.1.4.17 1.6.99.5)
Length=1670
Score = 108 bits (269), Expect = 1e-23, Method: Composition-based stats.
Identities = 52/161 (32%), Positives = 89/161 (55%), Gaps = 24/161 (14%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
D+ HF+ +++FR++ + L+ ++SE+ D W K +
Sbjct 1450 DMKNHFETVSRFRVRRNALDFDLSSEE-----------------------DFWFAVKIIM 1486
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCD-EAATDVPKSQIGFL 119
+CAD+ H +V W QH++W Q L EF+ QG +E+ L +SP+CD E ++V KSQ+GF+
Sbjct 1487 KCADLSHCSVPWSQHFQWCQRLSVEFYDQGDEEVARHLPMSPLCDREKHSEVAKSQLGFM 1546
Query 120 KLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQRISDS 160
+ PLFE L D +G++ C+S +K N+ W+ +S +
Sbjct 1547 SFVAVPLFEELMAIDGTGNIEKYCISVMKTNASHWEALSSA 1587
> tgo:TGME49_118680 3',5'--cyclic-nucleotide phosphodiesterase,
putative (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=812
Score = 98.2 bits (243), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 54/166 (32%), Positives = 84/166 (50%), Gaps = 27/166 (16%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
D+ THFDFL++FR + + N HK + D W A+ CI
Sbjct 486 DMKTHFDFLSRFR------------------NRRQAPGFNFHKVAE----DQWLAAEICI 523
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVP---KSQIG 117
R +D+GH+ +W+QH++W+ ++ EF+ QG +E LGL VSP+CD D P K Q G
Sbjct 524 RASDLGHAMTEWDQHFEWASRVVTEFYLQGEEERRLGLHVSPLCDR--DDHPSFSKCQHG 581
Query 118 FLKLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQRISDSEMD 163
FL + PL L + D + + LK+N E W+ ++ ++
Sbjct 582 FLGYVVKPLIAELGECDTLQRIIPQLMQNLKENMERWEAFTNEGVN 627
> tgo:TGME49_024840 cAMP phosphodiesterase, putative (EC:3.1.4.17
1.1.99.3)
Length=1324
Score = 95.9 bits (237), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 49/161 (30%), Positives = 85/161 (52%), Gaps = 23/161 (14%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
D+ HF+ L++FR++ + +P + D W +A+ C+
Sbjct 856 DMKQHFESLSRFRIRRN----------------------SPEFNYVKNVEDRWFVARMCV 893
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCD-EAATDVPKSQIGFL 119
+ AD+GHS V W QH++WS ++ EF+ QG +E + VSP+CD + D+ KSQ GFL
Sbjct 894 KVADIGHSCVPWNQHFEWSSRVVEEFYLQGDEERSRNMPVSPLCDRDKHADMAKSQGGFL 953
Query 120 KLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQRISDS 160
+ + PL + + + D G + + LS ++ N + WQ + +S
Sbjct 954 EFVVKPLIKEIDEIDPFGRVRSDILSHIEYNEKKWQELQES 994
> pfa:MAL13P1.118 3',5'-cyclic nucleotide phosphodiesterase (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=1139
Score = 95.1 bits (235), Expect = 1e-19, Method: Composition-based stats.
Identities = 52/162 (32%), Positives = 81/162 (50%), Gaps = 23/162 (14%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
D+ HF+ +++FR++ N + K S+ D L K I
Sbjct 959 DMKHHFEIISKFRIRRE--NEDFDYIKNSD--------------------DLLILTKMII 996
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAA-TDVPKSQIGFL 119
+ AD+ H +V W +HY W Q +++EF+ QG +EL+ + +SP+CD +V KSQI FL
Sbjct 997 KSADISHGSVSWSEHYCWCQRVLSEFYTQGDEELKNKMPLSPLCDRTKHNEVCKSQITFL 1056
Query 120 KLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQRISDSE 161
K + PLFE L+ DN+ + + CL L N W + E
Sbjct 1057 KFVVMPLFEELSHIDNNKFIKSFCLKRLNSNCIMWDTLMKEE 1098
> tgo:TGME49_041880 3', 5'-cyclic nucleotide phosphodiesterase,
putative (EC:2.1.1.43 3.1.4.17)
Length=1281
Score = 94.7 bits (234), Expect = 2e-19, Method: Composition-based stats.
Identities = 50/168 (29%), Positives = 80/168 (47%), Gaps = 24/168 (14%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
D+ HF+ +++FRL +++ DF +T+ D W L + CI
Sbjct 976 DMKMHFEMVSKFRL------------RRNSPDF-----------CRTNDDDVWMLLRMCI 1012
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAA-TDVPKSQIGFL 119
+ D+ H + W+ H WS +EF+ QG EL G ++P+ D D PK Q GFL
Sbjct 1013 KGGDLSHGLLSWDSHVSWSYRAASEFYEQGDLELAQGRTITPMFDRRKHADFPKGQEGFL 1072
Query 120 KLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQRISDSEMDWREI 167
+ I PL+E +A D + + CL NSE W+++ S ++ I
Sbjct 1073 RFIIIPLYEEIAAVDTTDVVQTCCLQNAVANSERWRQLQSSPEEFEAI 1120
> dre:564465 pde1a; phosphodiesterase 1A, calmodulin-dependent;
K13755 calcium/calmodulin-dependent 3',5'-cyclic nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=625
Score = 85.1 bits (209), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 37/72 (51%), Positives = 47/72 (65%), Gaps = 0/72 (0%)
Query 60 IRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFL 119
+ AD+ H A W+ HY+W+QALM EFF QG +E ELGL SP+CD AT + +SQIGF+
Sbjct 365 LHAADISHPAKGWKLHYRWTQALMEEFFRQGDKEAELGLPFSPLCDRKATMIAQSQIGFI 424
Query 120 KLICFPLFESLA 131
I P F L
Sbjct 425 DFIVEPTFSVLV 436
> tgo:TGME49_026770 cGMP-inhibited 3',5'-cyclic phosphodiesterase,
putative (EC:3.1.4.17)
Length=995
Score = 84.0 bits (206), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 44/132 (33%), Positives = 66/132 (50%), Gaps = 24/132 (18%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
D+S HF ++ R++ + +T N T+ D W + K C+
Sbjct 418 DMSQHFSTISTVRVR------------------RESRTFN----FITNEEDRWMMKKLCM 455
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVP--KSQIGF 118
+ D+GH+A+ W+QHYKWS + EF QG EL+ GL VSP+CD A ++ KSQ F
Sbjct 456 KIGDIGHAALDWDQHYKWSMRVTEEFLLQGDAELKAGLPVSPLCDRNAPEIELHKSQSSF 515
Query 119 LKLICFPLFESL 130
+ + PL L
Sbjct 516 INYLVVPLINEL 527
> hsa:5136 PDE1A, HCAM1, HSPDE1A, MGC26303; phosphodiesterase
1A, calmodulin-dependent (EC:3.1.4.17); K13755 calcium/calmodulin-dependent
3',5'-cyclic nucleotide phosphodiesterase [EC:3.1.4.17]
Length=535
Score = 82.8 bits (203), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 36/75 (48%), Positives = 47/75 (62%), Gaps = 0/75 (0%)
Query 59 CIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGF 118
+ AD+ H A W+ HY+W+ ALM EFF QG +E ELGL SP+CD +T V +SQIGF
Sbjct 361 ILHAADISHPAKSWKLHYRWTMALMEEFFLQGDKEAELGLPFSPLCDRKSTMVAQSQIGF 420
Query 119 LKLICFPLFESLAQA 133
+ I P F L +
Sbjct 421 IDFIVEPTFSLLTDS 435
> mmu:18573 Pde1a, AI987702, AW125737, MGC116577; phosphodiesterase
1A, calmodulin-dependent (EC:3.1.4.17); K13755 calcium/calmodulin-dependent
3',5'-cyclic nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=545
Score = 82.4 bits (202), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 36/75 (48%), Positives = 47/75 (62%), Gaps = 0/75 (0%)
Query 59 CIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGF 118
+ AD+ H A W+ HY+W+ ALM EFF QG +E ELGL SP+CD +T V +SQIGF
Sbjct 361 ILHAADISHPAKTWKLHYRWTMALMEEFFLQGDKEAELGLPFSPLCDRKSTMVAQSQIGF 420
Query 119 LKLICFPLFESLAQA 133
+ I P F L +
Sbjct 421 IDFIVEPTFSLLTDS 435
> mmu:18574 Pde1b, 63kDa, Pde1b1; phosphodiesterase 1B, Ca2+-calmodulin
dependent (EC:3.1.4.17); K13755 calcium/calmodulin-dependent
3',5'-cyclic nucleotide phosphodiesterase [EC:3.1.4.17]
Length=535
Score = 78.2 bits (191), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 34/72 (47%), Positives = 45/72 (62%), Gaps = 0/72 (0%)
Query 60 IRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFL 119
+ AD+ H +W H +W++ALM EFF QG +E ELGL SP+CD +T V +SQIGF+
Sbjct 365 LHAADISHPTKQWSVHSRWTKALMEEFFRQGDKEAELGLPFSPLCDRTSTLVAQSQIGFI 424
Query 120 KLICFPLFESLA 131
I P F L
Sbjct 425 DFIVEPTFSVLT 436
> tpv:TP01_1200 hypothetical protein
Length=840
Score = 77.4 bits (189), Expect = 2e-14, Method: Composition-based stats.
Identities = 42/132 (31%), Positives = 65/132 (49%), Gaps = 26/132 (19%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
DL HF + QFR++ + + EID T+ K I
Sbjct 717 DLEDHFQTITQFRVR----RASREFSVEGEID---------------------TICKMLI 751
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCD-EAATDVPKSQIGFL 119
+ +D+G S ++W +WSQ L+ EF+ QG +EL LGL ++P+CD +PK+Q GFL
Sbjct 752 KASDIGSSCMEWRLTLEWSQRLVQEFYTQGLEELSLGLPITPLCDPNKHNHLPKAQSGFL 811
Query 120 KLICFPLFESLA 131
+ PL+ +A
Sbjct 812 AIFVIPLYTEIA 823
> mmu:18575 Pde1c; phosphodiesterase 1C (EC:3.1.4.17); K13755
calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=631
Score = 77.4 bits (189), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 37/93 (39%), Positives = 53/93 (56%), Gaps = 6/93 (6%)
Query 59 CIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGF 118
+ AD+ H A W+ H++W+ +L+ EFF QG +E ELGL SP+CD +T V +SQ+GF
Sbjct 371 MLHTADISHPAKAWDLHHRWTMSLLEEFFRQGDREAELGLPFSPLCDRKSTMVAQSQVGF 430
Query 119 LKLICFPLFESLAQADNSGDLAAICLSFLKDNS 151
+ I P F L D+ +S L D S
Sbjct 431 IDFIVEPTFTVLT------DMTEKIVSPLIDES 457
> hsa:5137 PDE1C, Hcam3; phosphodiesterase 1C, calmodulin-dependent
70kDa (EC:3.1.4.17); K13755 calcium/calmodulin-dependent
3',5'-cyclic nucleotide phosphodiesterase [EC:3.1.4.17]
Length=634
Score = 77.4 bits (189), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 32/72 (44%), Positives = 46/72 (63%), Gaps = 0/72 (0%)
Query 59 CIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGF 118
+ AD+ H A W+ H++W+ +L+ EFF QG +E ELGL SP+CD +T V +SQ+GF
Sbjct 371 MLHTADISHPAKAWDLHHRWTMSLLEEFFRQGDREAELGLPFSPLCDRKSTMVAQSQVGF 430
Query 119 LKLICFPLFESL 130
+ I P F L
Sbjct 431 IDFIVEPTFTVL 442
> pfa:PFL0475w PDE1; cGMP-specific phosphodiesterase (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=954
Score = 77.0 bits (188), Expect = 3e-14, Method: Composition-based stats.
Identities = 42/156 (26%), Positives = 73/156 (46%), Gaps = 24/156 (15%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
D+ HF++++ FR F++ + L D W + +
Sbjct 792 DMKNHFEYISDFRTSKEFID---------------------YDNLSND--QIWQIFCLIL 828
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCD-EAATDVPKSQIGFL 119
+ +D+GHS ++W +H +W+ + EF+ QG E L +Q S +CD + SQI FL
Sbjct 829 KASDIGHSTLEWNKHLEWTLKINEEFYLQGLLEKSLNIQNSFLCDINTMNKLALSQIDFL 888
Query 120 KLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQ 155
K +C PLF L + D+ C+ +++N E W+
Sbjct 889 KHLCIPLFNELNYICKNNDVYTHCIQPIENNIERWE 924
> hsa:5153 PDE1B, PDE1B1, PDES1B; phosphodiesterase 1B, calmodulin-dependent
(EC:3.1.4.17); K13755 calcium/calmodulin-dependent
3',5'-cyclic nucleotide phosphodiesterase [EC:3.1.4.17]
Length=536
Score = 77.0 bits (188), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 34/72 (47%), Positives = 45/72 (62%), Gaps = 0/72 (0%)
Query 60 IRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFL 119
+ AD+ H +W H +W++ALM EFF QG +E ELGL SP+CD +T V +SQIGF+
Sbjct 366 LHAADISHPTKQWLVHSRWTKALMEEFFRQGDKEAELGLPFSPLCDRTSTLVAQSQIGFI 425
Query 120 KLICFPLFESLA 131
I P F L
Sbjct 426 DFIVEPTFSVLT 437
> tgo:TGME49_057080 cAMP-specific 3',5'-cyclic phosphodiesterase,
putative (EC:3.1.4.17)
Length=1691
Score = 77.0 bits (188), Expect = 4e-14, Method: Composition-based stats.
Identities = 46/166 (27%), Positives = 77/166 (46%), Gaps = 30/166 (18%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
D+S H ++++ R++ N VN+E D W L + CI
Sbjct 1460 DMSKHISYVSRLRVRAESGNFDVNNE-----------------------GDRWLLFQGCI 1496
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATD-VPKSQIGFL 119
+ AD+ H+A W+ H +W++ L EFF QG +E G+ V + D D P+SQ F+
Sbjct 1497 KAADLAHTATFWDNHKRWAECLCEEFFKQGDEERRQGMNVMDIFDRRQKDRFPQSQYRFI 1556
Query 120 KLICFPLFESLAQADN-----SGDLAAICLSFLKDNSETWQRISDS 160
+L+ PLF S+ ++ G IC + L N W+ +++
Sbjct 1557 ELVVEPLFNSVCSIEDLLKGRGGVRGKICKT-LSSNLARWKEAAEA 1601
> tgo:TGME49_080410 3',5'-cyclic phosphodiesterase, putative (EC:3.1.4.17
1.6.5.3); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=1085
Score = 76.6 bits (187), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 50/164 (30%), Positives = 77/164 (46%), Gaps = 30/164 (18%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
D+ THF+ +A FR++ +K E D +N + D + I
Sbjct 925 DMKTHFEQIANFRVR----------RQKEEFD-----PINNYD-------DRQKVMSMII 962
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVP--KSQIGF 118
+ AD+GH + W H +W ++ EF+ QG +E LGL VS +CD D KSQ+GF
Sbjct 963 KSADIGHGTLPWADHERWCNLVVQEFYEQGDEEKRLGLPVSFLCDRDQHDREFFKSQVGF 1022
Query 119 LKLICFPLFESLAQADNSGDLAA------ICLSFLKDNSETWQR 156
L + PL+E L + +L ICL L++N W++
Sbjct 1023 LDFVVKPLYEELKALETQLNLNPQLPIENICLKNLQENISEWKK 1066
> tgo:TGME49_066920 phosphodiesterase, putative (EC:3.1.4.17)
Length=1065
Score = 76.3 bits (186), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 42/158 (26%), Positives = 81/158 (51%), Gaps = 23/158 (14%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
D+ HF+ +++FRL + S DF + ++ D W + K +
Sbjct 726 DMKQHFETISRFRL------------RTSADDFDVAKHVD----------DRWAVLKMFV 763
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAA-TDVPKSQIGFL 119
+ AD+ H A+ W+ H++ S ++ EF+ QG +EL G+ +S +CD + ++PKSQ GF+
Sbjct 764 KMADISHVALPWDMHFRLSCDIVEEFYQQGDEELRRGMPLSQLCDRSKHNEMPKSQEGFI 823
Query 120 KLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQRI 157
+ + PL L + D++G + ++ N W+++
Sbjct 824 EFLAKPLVGVLIEGDSTGTIKTEVSEQMERNLLRWKQM 861
> tgo:TGME49_110520 calcium/calmodulin-dependent 3', 5'-cyclic
nucleotide phosphodiesterase, putative (EC:3.4.21.69 3.1.4.17)
Length=1531
Score = 75.9 bits (185), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 44/131 (33%), Positives = 67/131 (51%), Gaps = 23/131 (17%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
D+ HF+ +++FRL+ + + + N KK E D W + K
Sbjct 1330 DIKQHFETISRFRLRRN--SPEFNFLKKEEDD--------------------WLVRKMIF 1367
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCD-EAATDVPKSQIGFL 119
+ AD+ H+ V W+ H+ WS + EF+AQG E+ LGL VSP+CD E ++ KSQ+ FL
Sbjct 1368 KIADISHATVAWDAHFFWSCKVNAEFYAQGDAEVRLGLPVSPLCDREKHFEMGKSQVAFL 1427
Query 120 KLICFPLFESL 130
+ PL L
Sbjct 1428 NFVVEPLLREL 1438
> tgo:TGME49_072650 calcium/calmodulin-dependent 3',5'-cyclic
nucleotide phosphodiesterase, putative (EC:3.1.4.17)
Length=1320
Score = 75.5 bits (184), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 54/164 (32%), Positives = 75/164 (45%), Gaps = 24/164 (14%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
D+S HF+FL R+ N N + E D A +LA I
Sbjct 1175 DMSLHFEFLT--RVAARRENPDFNFVDREE-----------------DRALVLSLA---I 1212
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEA-ATDVPKSQIGFL 119
+ AD+GH A+ WE H WS L EF AQG +EL LGL VSP+CD + +SQ F+
Sbjct 1213 KVADLGHCALDWEAHTLWSARLKEEFLAQGDEELRLGLPVSPLCDRTQQGQLARSQANFI 1272
Query 120 KLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQRISDSEMD 163
+ + PL + LA A ++ + N W+ D E+D
Sbjct 1273 EFLFLPLAQHLAAVAPDDRFEATVVTRARQNISMWEGAQD-EVD 1315
> dre:563432 pde3b; phosphodiesterase 3B, cGMP-inhibited; K13296
cGMP-inhibited 3',5'-cyclic phosphodiesterase [EC:3.1.4.17]
Length=1107
Score = 75.1 bits (183), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 47/133 (35%), Positives = 64/133 (48%), Gaps = 20/133 (15%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
DL HFDFLA+F NSKVN ID+ T+ D + + CI
Sbjct 883 DLKKHFDFLAEF-------NSKVNDVNSPGIDW-------------TNENDRLLVCQVCI 922
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFLK 120
+ AD+ A H KW++ ++NEF+ QG +E LGL +SP D +A + K Q F+
Sbjct 923 KLADINGPAKDRSLHLKWTEGIVNEFYEQGDEEGTLGLPISPFMDRSAPQLAKLQESFIT 982
Query 121 LICFPLFESLAQA 133
I PL S A
Sbjct 983 HIVGPLCNSYDAA 995
> xla:496104 pde3b; phosphodiesterase 3b, cGMP-inhibited; K13296
cGMP-inhibited 3',5'-cyclic phosphodiesterase [EC:3.1.4.17]
Length=985
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 44/126 (34%), Positives = 63/126 (50%), Gaps = 20/126 (15%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
DL HFDFLA+F N+KVN ID+ ++ D + + CI
Sbjct 763 DLKKHFDFLAEF-------NAKVNDVNGLGIDW-------------SNENDRLLVCQVCI 802
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFLK 120
+ AD+ A E H KW++ ++NEF+ QG +E LGL +SP D +A + K Q F+
Sbjct 803 KLADINGPAKTRELHLKWTEGIVNEFYEQGDEESSLGLPISPFMDRSAPQLAKLQESFIT 862
Query 121 LICFPL 126
I PL
Sbjct 863 HIVGPL 868
> xla:444562 pde4b, MGC83972; phosphodiesterase 4B, cAMP-specific;
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=721
Score = 73.9 bits (180), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 38/111 (34%), Positives = 60/111 (54%), Gaps = 8/111 (7%)
Query 50 ADCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAAT 109
D + + + CAD+ + E + +W+ +M EFF QG +E E G+++SP+CD+
Sbjct 536 TDRIQVLRNMVHCADLSNPTKSLELYRQWTDRIMEEFFQQGDRERERGMEISPMCDKHTA 595
Query 110 DVPKSQIGFLKLICFPLFES---LAQADNSGDLAAICLSFLKDNSETWQRI 157
V KSQ+GF+ I PL+E+ L Q D A L L+DN +Q +
Sbjct 596 SVEKSQVGFIDYIVHPLWETWGDLVQPD-----AQDILDTLEDNRNWYQSM 641
> hsa:5142 PDE4B, DKFZp686F2182, DPDE4, MGC126529, PDE4B5, PDEIVB;
phosphodiesterase 4B, cAMP-specific (EC:3.1.4.17); K01120
3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=564
Score = 73.2 bits (178), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 38/111 (34%), Positives = 60/111 (54%), Gaps = 8/111 (7%)
Query 50 ADCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAAT 109
D + + + CAD+ + E + +W+ +M EFF QG +E E G+++SP+CD+
Sbjct 378 TDRIQVLRNMVHCADLSNPTKSLELYRQWTDRIMEEFFQQGDKERERGMEISPMCDKHTA 437
Query 110 DVPKSQIGFLKLICFPLFES---LAQADNSGDLAAICLSFLKDNSETWQRI 157
V KSQ+GF+ I PL+E+ L Q D A L L+DN +Q +
Sbjct 438 SVEKSQVGFIDYIVHPLWETWADLVQPD-----AQDILDTLEDNRNWYQSM 483
> mmu:18578 Pde4b, Dpde4, R74983, dunce; phosphodiesterase 4B,
cAMP specific (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=564
Score = 72.8 bits (177), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 38/111 (34%), Positives = 60/111 (54%), Gaps = 8/111 (7%)
Query 50 ADCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAAT 109
D + + + CAD+ + E + +W+ +M EFF QG +E E G+++SP+CD+
Sbjct 378 TDRIQVLRNMVHCADLSNPTKSLELYRQWTDRIMEEFFQQGDKERERGMEISPMCDKHTA 437
Query 110 DVPKSQIGFLKLICFPLFES---LAQADNSGDLAAICLSFLKDNSETWQRI 157
V KSQ+GF+ I PL+E+ L Q D A L L+DN +Q +
Sbjct 438 SVEKSQVGFIDYIVHPLWETWADLVQPD-----AQDILDTLEDNRNWYQSM 483
> dre:566998 novel protein similar to vertebrate phosphodiesterase
4D, cAMP-specific (phosphodiesterase E3 dunce homolog,
Drosophila) (PDE4D); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=745
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 37/106 (34%), Positives = 61/106 (57%), Gaps = 2/106 (1%)
Query 50 ADCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAAT 109
+D + + + CAD+ + + + +W+ +M EFF+QG +E E G+++SP+CD+
Sbjct 518 SDRIQVLQNMVHCADLSNPTKPLQLYRQWTDRIMEEFFSQGDRERERGMEISPMCDKHNA 577
Query 110 DVPKSQIGFLKLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQ 155
V KSQ+GF+ I PL+E+ AD A L L+DN E +Q
Sbjct 578 SVEKSQVGFIDYIVHPLWETW--ADLVHPDAQDILDTLEDNREWYQ 621
> dre:565706 cAMP-specific 3,5-cyclic phosphodiesterase 4B-like;
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=713
Score = 71.2 bits (173), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 36/108 (33%), Positives = 59/108 (54%), Gaps = 2/108 (1%)
Query 50 ADCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAAT 109
D + + + CAD+ + E + +W+ +M EFF QG +E E G+++SP+CD+
Sbjct 528 TDRIQVLRNMVHCADLSNPTKSLELYRQWTDRIMEEFFHQGDRERERGMEISPMCDKHTA 587
Query 110 DVPKSQIGFLKLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQRI 157
V KSQ+GF+ I PL+E+ A + A L L+DN +Q +
Sbjct 588 SVEKSQVGFIDYIVHPLWETWADLVHPD--AQDILDTLEDNRNWYQSM 633
> dre:572061 pde4a; phosphodiesterase 4A, cAMP-specific; K01120
3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=736
Score = 71.2 bits (173), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 37/109 (33%), Positives = 60/109 (55%), Gaps = 10/109 (9%)
Query 51 DCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATD 110
D + + + CAD+ + + +W++ +M EFF QG +E E G+++SP+CD+
Sbjct 537 DRIQVLRNMVHCADLSNPTKPLAVYRQWTERIMEEFFQQGDKERERGMEISPMCDKHTAS 596
Query 111 VPKSQIGFLKLICFPLFESLAQADNSGDL----AAICLSFLKDNSETWQ 155
V KSQ+GF+ I PL+E+ GDL A L L+DN + +Q
Sbjct 597 VEKSQVGFIDYIVHPLWETW------GDLVHPDAQEILDTLEDNRDWYQ 639
> mmu:238871 Pde4d, 9630011N22Rik, Dpde3; phosphodiesterase 4D,
cAMP specific (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=747
Score = 70.5 bits (171), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 36/106 (33%), Positives = 60/106 (56%), Gaps = 2/106 (1%)
Query 50 ADCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAAT 109
+D + + + CAD+ + + + +W+ +M EFF QG +E E G+++SP+CD+
Sbjct 545 SDRIQVLQNMVHCADLSNPTKPLQLYRQWTDRIMEEFFRQGDRERERGMEISPMCDKHNA 604
Query 110 DVPKSQIGFLKLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQ 155
V KSQ+GF+ I PL+E+ A + A L L+DN E +Q
Sbjct 605 SVEKSQVGFIDYIVHPLWETWADLVHPD--AQDILDTLEDNREWYQ 648
> hsa:5140 PDE3B, HcGIP1, cGIPDE1; phosphodiesterase 3B, cGMP-inhibited
(EC:3.1.4.17); K13296 cGMP-inhibited 3',5'-cyclic
phosphodiesterase [EC:3.1.4.17]
Length=1112
Score = 70.5 bits (171), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 52/175 (29%), Positives = 81/175 (46%), Gaps = 24/175 (13%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
DL HFDFLA+F N+K N + I++ ++ D + + CI
Sbjct 894 DLKKHFDFLAEF-------NAKANDVNSNGIEW-------------SNENDRLLVCQVCI 933
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFLK 120
+ AD+ A + H KW++ ++NEF+ QG +E LGL +SP D ++ + K Q F+
Sbjct 934 KLADINGPAKVRDLHLKWTEGIVNEFYEQGDEEANLGLPISPFMDRSSPQLAKLQESFIT 993
Query 121 LICFPLFESLAQADNSGDLAAICLSFLKDN-SETWQRISDSEMDWREISQINKCN 174
I PL S D +G L L +DN +E+ E+D + N N
Sbjct 994 HIVGPLCNSY---DAAGLLPGQWLEAEEDNDTESGDDEDGEELDTEDEEMENNLN 1045
> hsa:5144 PDE4D, DKFZp686M11213, DPDE3, FLJ97311, HSPDE4D, PDE4DN2,
STRK1; phosphodiesterase 4D, cAMP-specific (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=809
Score = 70.5 bits (171), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 37/106 (34%), Positives = 60/106 (56%), Gaps = 2/106 (1%)
Query 50 ADCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAAT 109
+D + + + CAD+ + + + +W+ +M EFF QG +E E G+++SP+CD+
Sbjct 606 SDRIQVLQNMVHCADLSNPTKPLQLYRQWTDRIMEEFFRQGDRERERGMEISPMCDKHNA 665
Query 110 DVPKSQIGFLKLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQ 155
V KSQ+GF+ I PL+E+ AD A L L+DN E +Q
Sbjct 666 SVEKSQVGFIDYIVHPLWETW--ADLVHPDAQDILDTLEDNREWYQ 709
> dre:571344 fc68c05, si:ch211-255d18.8, wu:fc68c05; si:ch211-255d18.1
Length=709
Score = 70.1 bits (170), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 46/157 (29%), Positives = 76/157 (48%), Gaps = 19/157 (12%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
D+S H +FLA + + KV S+ +D S D + + +
Sbjct 472 DMSKHMNFLADLKTMVE--TKKVTSQGVLLLDNYS---------------DRIQVLQNMV 514
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFLK 120
CAD+ + E + KW+ +M EFF+QG +E + G+ VSP+CD+ + +Q+GF+
Sbjct 515 HCADLSNPTKPLELYRKWTDRIMVEFFSQGDRERDKGIDVSPMCDKHTASMENTQVGFID 574
Query 121 LICFPLFESLAQADNSGDLAAICLSFLKDNSETWQRI 157
I PL+E+ A + A L L+DN E +Q +
Sbjct 575 YIIHPLWETWADLVHPD--AQDILDTLEDNREWYQSM 609
> xla:495146 hypothetical LOC495146; K01120 3',5'-cyclic-nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=682
Score = 70.1 bits (170), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 36/109 (33%), Positives = 59/109 (54%), Gaps = 8/109 (7%)
Query 50 ADCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAAT 109
D + + + CAD+ + E + +W+ ++ EFF QG +E E G+++SP+CD+
Sbjct 488 TDRIQVLRNMVHCADLSNPTKPLELYRQWTDRILEEFFRQGDKERERGMEISPMCDKHTA 547
Query 110 DVPKSQIGFLKLICFPLFES---LAQADNSGDLAAICLSFLKDNSETWQ 155
V KSQ+GF+ I PL+E+ L D A L L+DN + +Q
Sbjct 548 SVEKSQVGFIDYIVHPLWETWSDLVHPD-----AQDILDTLEDNRDWYQ 591
> dre:565259 pde4c, im:7160317, si:dkey-149i17.5; phosphodiesterase
4C, cAMP-specific (phosphodiesterase E1 dunce homolog,
Drosophila)
Length=672
Score = 69.7 bits (169), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 36/111 (32%), Positives = 61/111 (54%), Gaps = 8/111 (7%)
Query 50 ADCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAAT 109
+D + + + CAD+ + E + +W+ +M EFF QG +E + G+++SP+CD+
Sbjct 518 SDRIQVLQNMVHCADLSNPTKPLELYRQWTDRIMVEFFTQGDRERDKGMEISPMCDKHNA 577
Query 110 DVPKSQIGFLKLICFPLFES---LAQADNSGDLAAICLSFLKDNSETWQRI 157
+ KSQ+GF+ I PL+E+ L D A L L+DN E +Q +
Sbjct 578 SIEKSQVGFIDYIVHPLWETWADLVHPD-----AQEILDTLEDNREWYQSM 623
> cel:R153.1 pde-4; PhosphoDiEsterase family member (pde-4); K01120
3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=673
Score = 69.3 bits (168), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 30/81 (37%), Positives = 50/81 (61%), Gaps = 0/81 (0%)
Query 51 DCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATD 110
D + ++ I AD+ + E + +W+Q +M E++ QG +E ELGL++SP+CD
Sbjct 551 DKIQVLQSMIHLADLSNPTKPIELYQQWNQRIMEEYWRQGDKEKELGLEISPMCDRGNVT 610
Query 111 VPKSQIGFLKLICFPLFESLA 131
+ KSQ+GF+ I PL+E+ A
Sbjct 611 IEKSQVGFIDYIVHPLYETWA 631
> mmu:18576 Pde3b, 9830102A01Rik, AI847709; phosphodiesterase
3B, cGMP-inhibited (EC:3.1.4.17); K13296 cGMP-inhibited 3',5'-cyclic
phosphodiesterase [EC:3.1.4.17]
Length=1099
Score = 69.3 bits (168), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 42/133 (31%), Positives = 64/133 (48%), Gaps = 20/133 (15%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
DL HFDFLA+F N+K N + I++ S D + + CI
Sbjct 870 DLKKHFDFLAEF-------NAKANDVNSNGIEWSSEN-------------DRLLVCQVCI 909
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFLK 120
+ AD+ A + H +W++ ++NEF+ QG +E LGL +SP D ++ + K Q F+
Sbjct 910 KLADINGPAKDRDLHLRWTEGIVNEFYEQGDEEATLGLPISPFMDRSSPQLAKLQESFIT 969
Query 121 LICFPLFESLAQA 133
I PL S A
Sbjct 970 HIVGPLCNSYDAA 982
> dre:567051 si:dkey-48h7.2; K13296 cGMP-inhibited 3',5'-cyclic
phosphodiesterase [EC:3.1.4.17]
Length=1117
Score = 69.3 bits (168), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 44/133 (33%), Positives = 65/133 (48%), Gaps = 20/133 (15%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
DL HFDFLA+F N+KV + S ID+ T+ D + + CI
Sbjct 835 DLKKHFDFLAEF-------NAKVGDDGCSGIDW-------------TNENDRLLVCQMCI 874
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFLK 120
+ AD+ + H +W++ ++NEF+ QG +E LGL +SP D +A + K Q F+
Sbjct 875 KLADINGPLKCKDLHLQWTEGIVNEFYEQGDEESSLGLPISPFMDRSAPQLAKLQESFIT 934
Query 121 LICFPLFESLAQA 133
I PL S A
Sbjct 935 HIVGPLCSSYDSA 947
> hsa:5141 PDE4A, DPDE2, PDE4, PDE46; phosphodiesterase 4A, cAMP-specific
(EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=886
Score = 69.3 bits (168), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 30/82 (36%), Positives = 50/82 (60%), Gaps = 0/82 (0%)
Query 50 ADCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAAT 109
+D + + + CAD+ + E + +W+ +M EFF QG +E E G+++SP+CD+
Sbjct 577 SDRIQVLRNMVHCADLSNPTKPLELYRQWTDRIMAEFFQQGDRERERGMEISPMCDKHTA 636
Query 110 DVPKSQIGFLKLICFPLFESLA 131
V KSQ+GF+ I PL+E+ A
Sbjct 637 SVEKSQVGFIDYIVHPLWETWA 658
> hsa:27115 PDE7B, MGC88256, bA472E5.1; phosphodiesterase 7B (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=450
Score = 68.9 bits (167), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 55/110 (50%), Gaps = 0/110 (0%)
Query 48 DPADCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEA 107
D D + + ++CAD+ + WE +WS+ + EF+ QG E + L++SP+C++
Sbjct 307 DAQDRHFMLQIALKCADICNPCRIWEMSKQWSERVCEEFYRQGELEQKFELEISPLCNQQ 366
Query 108 ATDVPKSQIGFLKLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQRI 157
+P QIGF+ I PLF A + L+ L L N W+ +
Sbjct 367 KDSIPSIQIGFMSYIVEPLFREWAHFTGNSTLSENMLGHLAHNKAQWKSL 416
> mmu:29863 Pde7b; phosphodiesterase 7B (EC:3.1.4.17); K01120
3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=446
Score = 68.9 bits (167), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 34/115 (29%), Positives = 59/115 (51%), Gaps = 0/115 (0%)
Query 55 LAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKS 114
+ + ++CAD+ + WE +WS+ + EF+ QG E + L++SP+C++ +P
Sbjct 314 MLQIALKCADICNPCRIWEMSKQWSERVCEEFYRQGDLEQKFELEISPLCNQQKDSIPSI 373
Query 115 QIGFLKLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQRISDSEMDWREISQ 169
QIGF+ I PLF A+ + L+ LS L N W+ + ++ R Q
Sbjct 374 QIGFMTYIVEPLFREWARFTGNSTLSENMLSHLAHNKAQWKSLLSNQHRRRGSGQ 428
> mmu:18577 Pde4a, D9Ertd60e, Dpde2; phosphodiesterase 4A, cAMP
specific (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=610
Score = 68.9 bits (167), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 30/82 (36%), Positives = 50/82 (60%), Gaps = 0/82 (0%)
Query 50 ADCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAAT 109
+D + + + CAD+ + E + +W+ +M EFF QG +E E G+++SP+CD+
Sbjct 329 SDRIQVLRNMVHCADLSNPTKPLELYRQWTDRIMAEFFQQGDRERERGMEISPMCDKHTA 388
Query 110 DVPKSQIGFLKLICFPLFESLA 131
V KSQ+GF+ I PL+E+ A
Sbjct 389 SVEKSQVGFIDYIVHPLWETWA 410
> pfa:MAL13P1.119 calcium/calmodulin-dependent 3',5'-cyclic nucleotide
phosphodiesterase 1b (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=769
Score = 68.6 bits (166), Expect = 1e-11, Method: Composition-based stats.
Identities = 45/175 (25%), Positives = 81/175 (46%), Gaps = 27/175 (15%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
D+S H LAQFR++ + K+ S + I K I
Sbjct 600 DMSKHIKILAQFRIK----SIKIKSYIEKNIIL---------------------CLKMII 634
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAA-TDVPKSQIGFL 119
+ AD+ H+ V W +HY+W + L+NEF+ +G + ++G +++P+ D + + Q FL
Sbjct 635 KAADLSHNCVDWSEHYQWVKRLVNEFYYEGDELFQMGYKINPLFDRNCHNNFIQIQRTFL 694
Query 120 KLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQRISDSEMDWREISQINKCN 174
K + +PL SL DN+ + ++ +K N W +I ++ ++ CN
Sbjct 695 KELVYPLIISLKTLDNT-SITQDMINNVKRNYSKWTKIEKCQIKKKKYLNELLCN 748
> hsa:5143 PDE4C, DPDE1, MGC126222; phosphodiesterase 4C, cAMP-specific
(EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=712
Score = 68.6 bits (166), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 47/155 (30%), Positives = 75/155 (48%), Gaps = 19/155 (12%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
D+S H + LA L + V ++K + + L + +D + + +
Sbjct 500 DMSKHMNLLAD-------LKTMVETKKVTSLGV----------LLLDNYSDRIQVLQNLV 542
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFLK 120
CAD+ + + +W+ +M EFF QG +E E GL +SP+CD+ V KSQ+GF+
Sbjct 543 HCADLSNPTKPLPLYRQWTDRIMAEFFQQGDRERESGLDISPMCDKHTASVEKSQVGFID 602
Query 121 LICFPLFESLAQADNSGDLAAICLSFLKDNSETWQ 155
I PL+E+ AD A L L+DN E +Q
Sbjct 603 YIAHPLWETW--ADLVHPDAQDLLDTLEDNREWYQ 635
> hsa:5139 PDE3A, CGI-PDE; phosphodiesterase 3A, cGMP-inhibited
(EC:3.1.4.17); K13296 cGMP-inhibited 3',5'-cyclic phosphodiesterase
[EC:3.1.4.17]
Length=1141
Score = 65.9 bits (159), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 44/133 (33%), Positives = 63/133 (47%), Gaps = 22/133 (16%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
DL HFDF+A+F N KVN + ID+ T+ D + + CI
Sbjct 909 DLKKHFDFVAKF-------NGKVNDDVG--IDW-------------TNENDRLLVCQMCI 946
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFLK 120
+ AD+ A E H +W+ ++NEF+ QG +E LGL +SP D +A + Q F+
Sbjct 947 KLADINGPAKCKELHLQWTDGIVNEFYEQGDEEASLGLPISPFMDRSAPQLANLQESFIS 1006
Query 121 LICFPLFESLAQA 133
I PL S A
Sbjct 1007 HIVGPLCNSYDSA 1019
> dre:100151215 phosphodiesterase 7A-like (EC:3.1.4.17); K01120
3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=490
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 33/98 (33%), Positives = 53/98 (54%), Gaps = 1/98 (1%)
Query 57 KACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQI 116
+ ++CAD+ + WE +WS+ + EFF QG E +L L++SP+CD A + QI
Sbjct 357 QMALKCADICNPCRPWELSKQWSEKVTEEFFHQGDIEKKLKLEISPLCDSEANTIASVQI 416
Query 117 GFLKLICFPLFESLAQADNSGDLAAICLSFLKDNSETW 154
GF+ + PLF A+ ++ L+ L L N +W
Sbjct 417 GFMTYVVEPLFAEWARFSDT-RLSQTMLGHLGLNKASW 453
> mmu:54611 Pde3a, A930022O17Rik, C87899; phosphodiesterase 3A,
cGMP inhibited (EC:3.1.4.17); K13296 cGMP-inhibited 3',5'-cyclic
phosphodiesterase [EC:3.1.4.17]
Length=1141
Score = 64.7 bits (156), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 43/133 (32%), Positives = 64/133 (48%), Gaps = 22/133 (16%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
DL HFDF+A+F N+KVN + ID+ T+ D + + CI
Sbjct 909 DLKKHFDFVAKF-------NAKVNDDVG--IDW-------------TNENDRLLVCQMCI 946
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFLK 120
+ AD+ A E H +W++ + +EF+ QG +E LGL +SP D +A + Q F+
Sbjct 947 KLADINGPAKCKELHLRWTEGIASEFYEQGDEEASLGLPISPFMDRSAPQLANLQESFIS 1006
Query 121 LICFPLFESLAQA 133
I PL S A
Sbjct 1007 HIVGPLCHSYDSA 1019
> cel:T04D3.3 pde-1; PhosphoDiEsterase family member (pde-1);
K13755 calcium/calmodulin-dependent 3',5'-cyclic nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=664
Score = 63.9 bits (154), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 36/66 (54%), Gaps = 0/66 (0%)
Query 60 IRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFL 119
+ D+ H A W H +W++ ++ EFF QG E +GL SP+CD V SQIGF+
Sbjct 476 VHACDISHPAKPWNLHERWTEGVLEEFFRQGDLEASMGLPYSPLCDRHTVHVADSQIGFI 535
Query 120 KLICFP 125
I P
Sbjct 536 DFIVEP 541
> hsa:5150 PDE7A, HCP1, PDE7; phosphodiesterase 7A (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=456
Score = 62.0 bits (149), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 32/99 (32%), Positives = 51/99 (51%), Gaps = 1/99 (1%)
Query 57 KACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQI 116
+ ++CAD+ + WE +WS+ + EFF QG E + L VSP+CD + QI
Sbjct 329 QMALKCADICNPCRTWELSKQWSEKVTEEFFHQGDIEKKYHLGVSPLCDRHTESIANIQI 388
Query 117 GFLKLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQ 155
GF+ + PLF A+ N+ L+ L + N +W+
Sbjct 389 GFMTYLVEPLFTEWARFSNT-RLSQTMLGHVGLNKASWK 426
Lambda K H
0.320 0.132 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4976880524
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40