bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1288_orf1
Length=169
Score E
Sequences producing significant alignments: (Bits) Value
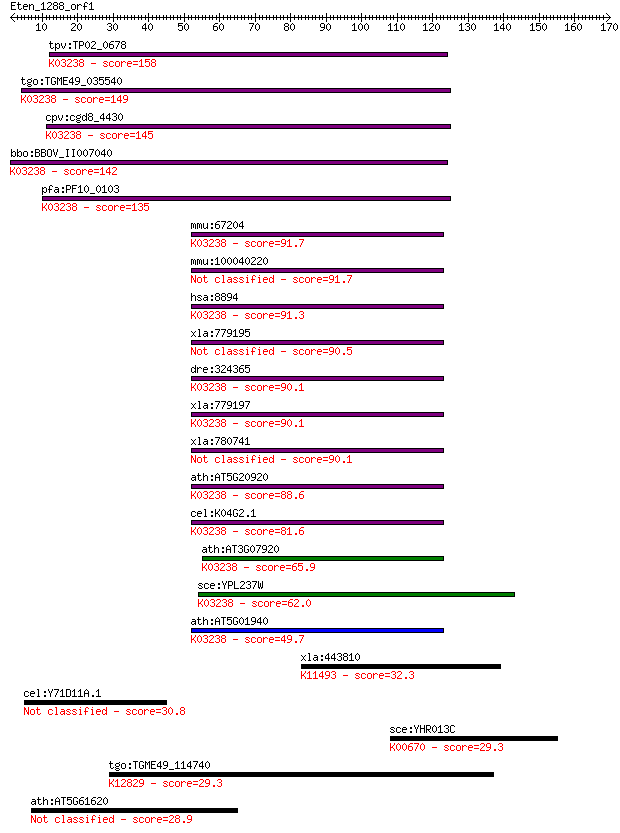
tpv:TP02_0678 eukaryotic translation initiation factor 2 subun... 158 9e-39
tgo:TGME49_035540 translation initiation factor 2 beta, putati... 149 3e-36
cpv:cgd8_4430 translation initiation factor if-2 betam beta su... 145 5e-35
bbo:BBOV_II007040 18.m06583; eukaryotic translation initiation... 142 4e-34
pfa:PF10_0103 eukaryotic translation initiation factor 2 beta,... 135 7e-32
mmu:67204 Eif2s2, 2810026E11Rik, 38kDa, AA408636, AA571381, AA... 91.7 1e-18
mmu:100040220 Gm9892; predicted gene 9892 91.7
hsa:8894 EIF2S2, DKFZp686L18198, EIF2, EIF2B, EIF2beta, MGC850... 91.3 1e-18
xla:779195 hypothetical protein MGC130708 90.5 2e-18
dre:324365 eif2s2, wu:fc26g03, wu:fu10f07, zgc:77084; eukaryot... 90.1 3e-18
xla:779197 eif2s2, MGC130959; eukaryotic translation initiatio... 90.1 4e-18
xla:780741 eukaryote initiation factor 2 beta 90.1 4e-18
ath:AT5G20920 EIF2 BETA; translation initiation factor; K03238... 88.6 9e-18
cel:K04G2.1 iftb-1; eIFTwoBeta (eIF2beta translation initiatio... 81.6 1e-15
ath:AT3G07920 translation initiation factor; K03238 translatio... 65.9 7e-11
sce:YPL237W SUI3; Beta subunit of the translation initiation f... 62.0 8e-10
ath:AT5G01940 eukaryotic translation initiation factor 2B fami... 49.7 4e-06
xla:443810 rcc1-b, MGC115703, MGC132085, RanGEF, chc1, chc1-b,... 32.3 0.76
cel:Y71D11A.1 cdh-12; CaDHerin family member (cdh-12) 30.8 2.4
sce:YHR013C ARD1, NAA10; Ard1p (EC:2.3.1.88); K00670 peptide a... 29.3 6.0
tgo:TGME49_114740 splicing factor 3B subunit 2, putative ; K12... 29.3 6.8
ath:AT5G61620 myb family transcription factor 28.9 8.0
> tpv:TP02_0678 eukaryotic translation initiation factor 2 subunit
beta; K03238 translation initiation factor 2 subunit 2
Length=241
Score = 158 bits (399), Expect = 9e-39, Method: Compositional matrix adjust.
Identities = 73/112 (65%), Positives = 88/112 (78%), Gaps = 5/112 (4%)
Query 12 FDFGERRKKKKEKKPEDQAAEAASEAFVDGSGQLFVRGHVYSYEEMLKRIQELIVRNNPD 71
F+FGE++KK+ A ++ +DG+GQ+FV+GHVY YEE+L RIQ +I NNPD
Sbjct 49 FNFGEKKKKRTV-----DAGDSPKTEIIDGTGQVFVKGHVYPYEELLARIQSMINANNPD 103
Query 72 LAGSKRYTIKPPQVVRVGSKKVAWINFKDICGIMHRPSEHVLQFVLAELGTE 123
L+GSKRYTIKPPQVVRVGSKKVAWINF+DIC IM R +HV QFVLAELGTE
Sbjct 104 LSGSKRYTIKPPQVVRVGSKKVAWINFRDICSIMGRSMDHVHQFVLAELGTE 155
> tgo:TGME49_035540 translation initiation factor 2 beta, putative
; K03238 translation initiation factor 2 subunit 2
Length=231
Score = 149 bits (377), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 83/122 (68%), Positives = 98/122 (80%), Gaps = 3/122 (2%)
Query 4 AEHRAAQAFDFGERRKKKKEKKPED-QAAEAASEAFVDGSGQLFVRGHVYSYEEMLKRIQ 62
E A Q FDFGERR++KK++K E QA E A +DG+G+LF RG YSY+EML+RIQ
Sbjct 26 GEEEAPQQFDFGERRRRKKKEKKETEQAQEEAP--IIDGTGELFKRGKEYSYQEMLQRIQ 83
Query 63 ELIVRNNPDLAGSKRYTIKPPQVVRVGSKKVAWINFKDICGIMHRPSEHVLQFVLAELGT 122
LI ++NPDL+G+KRYTIKPPQVVRVGSKKVAWINFKDIC IM+R +HV QFVLAELGT
Sbjct 84 TLIEKHNPDLSGAKRYTIKPPQVVRVGSKKVAWINFKDICNIMNRQPDHVHQFVLAELGT 143
Query 123 ES 124
E
Sbjct 144 EG 145
> cpv:cgd8_4430 translation initiation factor if-2 betam beta
subunit ZnR ; K03238 translation initiation factor 2 subunit
2
Length=214
Score = 145 bits (367), Expect = 5e-35, Method: Compositional matrix adjust.
Identities = 72/114 (63%), Positives = 95/114 (83%), Gaps = 1/114 (0%)
Query 11 AFDFGERRKKKKEKKPEDQAAEAASEAFVDGSGQLFVRGHVYSYEEMLKRIQELIVRNNP 70
+DFGE++KKKK K+ + +E S ++VDGSGQLFV+G +Y YEE+L+R++ LI+ +NP
Sbjct 16 GYDFGEKKKKKKSKEHKTGVSEVES-SYVDGSGQLFVKGAIYPYEELLERVRRLILEHNP 74
Query 71 DLAGSKRYTIKPPQVVRVGSKKVAWINFKDICGIMHRPSEHVLQFVLAELGTES 124
DL G+KRYT+KPPQVVRVGSKKVAWINF++IC IM R ++HV QFVL+ELGTE
Sbjct 75 DLWGAKRYTLKPPQVVRVGSKKVAWINFQEICNIMQRNADHVFQFVLSELGTEG 128
> bbo:BBOV_II007040 18.m06583; eukaryotic translation initiation
factor 2, beta; K03238 translation initiation factor 2 subunit
2
Length=238
Score = 142 bits (359), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 72/123 (58%), Positives = 91/123 (73%), Gaps = 5/123 (4%)
Query 1 AQPAEHRAAQAFDFGERRKKKKEKKPEDQAAEAASEAFVDGSGQLFVRGHVYSYEEMLKR 60
A + A +DFGE++KKK A+++ +DGSGQ+FVRGHVY YEE+L R
Sbjct 35 AAAVKAEGAPKYDFGEKKKKKAAP-----ASDSVKSEVIDGSGQVFVRGHVYQYEELLNR 89
Query 61 IQELIVRNNPDLAGSKRYTIKPPQVVRVGSKKVAWINFKDICGIMHRPSEHVLQFVLAEL 120
IQ LI ++ DL+G+K+YTI+PPQVVRVGSKKVAWINFK++C IM R +HV QFVLAEL
Sbjct 90 IQTLINAHSHDLSGNKKYTIRPPQVVRVGSKKVAWINFKELCNIMGRTMDHVHQFVLAEL 149
Query 121 GTE 123
GTE
Sbjct 150 GTE 152
> pfa:PF10_0103 eukaryotic translation initiation factor 2 beta,
putative; K03238 translation initiation factor 2 subunit
2
Length=222
Score = 135 bits (340), Expect = 7e-32, Method: Compositional matrix adjust.
Identities = 74/115 (64%), Positives = 89/115 (77%), Gaps = 4/115 (3%)
Query 10 QAFDFGERRKKKKEKKPEDQAAEAASEAFVDGSGQLFVRGHVYSYEEMLKRIQELIVRNN 69
Q FDFGE++KKKK+K + E E +DG+G++F RG VY Y+E+L RIQ+LI ++N
Sbjct 25 QLFDFGEKKKKKKKK----EVVEKVEEIIIDGTGKVFERGAVYPYDELLHRIQDLINKHN 80
Query 70 PDLAGSKRYTIKPPQVVRVGSKKVAWINFKDICGIMHRPSEHVLQFVLAELGTES 124
DL SK+YTIKPPQVVRVGSKKVAWINFKDIC IM+R EHV FVLAELGTE
Sbjct 81 IDLCISKKYTIKPPQVVRVGSKKVAWINFKDICTIMNRNEEHVFHFVLAELGTEG 135
> mmu:67204 Eif2s2, 2810026E11Rik, 38kDa, AA408636, AA571381,
AA986487, AW822225, D2Ertd303e, EIF2, EIF2B, MGC130606; eukaryotic
translation initiation factor 2, subunit 2 (beta); K03238
translation initiation factor 2 subunit 2
Length=331
Score = 91.7 bits (226), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 40/73 (54%), Positives = 57/73 (78%), Gaps = 2/73 (2%)
Query 52 YSYEEMLKRIQELIVRNNPDL-AGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPS 109
Y+YEE+L R+ ++ NPD+ AG KR + +KPPQVVRVG+KK +++NF DIC ++HR
Sbjct 172 YTYEELLNRVFNIMREKNPDMVAGEKRKFVMKPPQVVRVGTKKTSFVNFTDICKLLHRQP 231
Query 110 EHVLQFVLAELGT 122
+H+L F+LAELGT
Sbjct 232 KHLLAFLLAELGT 244
> mmu:100040220 Gm9892; predicted gene 9892
Length=331
Score = 91.7 bits (226), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 39/73 (53%), Positives = 57/73 (78%), Gaps = 2/73 (2%)
Query 52 YSYEEMLKRIQELIVRNNPDL-AGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPS 109
Y+YEE+L R+ ++ NPD+ AG KR + +KPPQV+RVG+KK +++NF DIC ++HR
Sbjct 172 YTYEELLNRVLNIMREKNPDMVAGEKRKFVMKPPQVIRVGTKKTSFVNFTDICKLLHRQP 231
Query 110 EHVLQFVLAELGT 122
+H+L F+LAELGT
Sbjct 232 KHLLAFLLAELGT 244
> hsa:8894 EIF2S2, DKFZp686L18198, EIF2, EIF2B, EIF2beta, MGC8508;
eukaryotic translation initiation factor 2, subunit 2 beta,
38kDa; K03238 translation initiation factor 2 subunit 2
Length=333
Score = 91.3 bits (225), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 40/73 (54%), Positives = 57/73 (78%), Gaps = 2/73 (2%)
Query 52 YSYEEMLKRIQELIVRNNPDL-AGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPS 109
Y+YEE+L R+ ++ NPD+ AG KR + +KPPQVVRVG+KK +++NF DIC ++HR
Sbjct 174 YTYEELLNRVFNIMREKNPDMVAGEKRKFVMKPPQVVRVGTKKTSFVNFTDICKLLHRQP 233
Query 110 EHVLQFVLAELGT 122
+H+L F+LAELGT
Sbjct 234 KHLLAFLLAELGT 246
> xla:779195 hypothetical protein MGC130708
Length=332
Score = 90.5 bits (223), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 39/73 (53%), Positives = 57/73 (78%), Gaps = 2/73 (2%)
Query 52 YSYEEMLKRIQELIVRNNPDL-AGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPS 109
Y+Y+E+L R+ ++ NPD+ AG KR + +KPPQVVRVG+KK +++NF DIC ++HR
Sbjct 173 YTYDELLNRVFNIMREKNPDMVAGEKRKFVMKPPQVVRVGTKKTSFVNFTDICKLLHRQP 232
Query 110 EHVLQFVLAELGT 122
+H+L F+LAELGT
Sbjct 233 KHLLAFLLAELGT 245
> dre:324365 eif2s2, wu:fc26g03, wu:fu10f07, zgc:77084; eukaryotic
translation initiation factor 2, subunit 2 beta; K03238
translation initiation factor 2 subunit 2
Length=327
Score = 90.1 bits (222), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 39/73 (53%), Positives = 57/73 (78%), Gaps = 2/73 (2%)
Query 52 YSYEEMLKRIQELIVRNNPDL-AGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPS 109
Y+Y+E+L R+ ++ NPD+ AG KR + +KPPQVVRVG+KK +++NF DIC ++HR
Sbjct 168 YTYDELLNRVFNIMREKNPDMVAGEKRKFVMKPPQVVRVGTKKTSFVNFTDICKLLHRQP 227
Query 110 EHVLQFVLAELGT 122
+H+L F+LAELGT
Sbjct 228 KHLLVFLLAELGT 240
> xla:779197 eif2s2, MGC130959; eukaryotic translation initiation
factor 2, subunit 2 beta, 38kDa; K03238 translation initiation
factor 2 subunit 2
Length=335
Score = 90.1 bits (222), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 39/73 (53%), Positives = 57/73 (78%), Gaps = 2/73 (2%)
Query 52 YSYEEMLKRIQELIVRNNPDL-AGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPS 109
Y+Y+E+L R+ ++ NPD+ AG KR + +KPPQVVRVG+KK +++NF DIC ++HR
Sbjct 176 YTYDELLNRVFNIMREKNPDMVAGEKRKFVMKPPQVVRVGTKKTSFVNFTDICKLLHRQP 235
Query 110 EHVLQFVLAELGT 122
+H+L F+LAELGT
Sbjct 236 KHLLAFLLAELGT 248
> xla:780741 eukaryote initiation factor 2 beta
Length=335
Score = 90.1 bits (222), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 39/73 (53%), Positives = 57/73 (78%), Gaps = 2/73 (2%)
Query 52 YSYEEMLKRIQELIVRNNPDL-AGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPS 109
Y+Y+E+L R+ ++ NPD+ AG KR + +KPPQVVRVG+KK +++NF DIC ++HR
Sbjct 176 YTYDELLNRVFNIMREKNPDMVAGEKRKFVMKPPQVVRVGTKKTSFVNFTDICKLLHRQP 235
Query 110 EHVLQFVLAELGT 122
+H+L F+LAELGT
Sbjct 236 KHLLAFLLAELGT 248
> ath:AT5G20920 EIF2 BETA; translation initiation factor; K03238
translation initiation factor 2 subunit 2
Length=268
Score = 88.6 bits (218), Expect = 9e-18, Method: Compositional matrix adjust.
Identities = 37/72 (51%), Positives = 55/72 (76%), Gaps = 1/72 (1%)
Query 52 YSYEEMLKRIQELIVRNNPDLAGSKRYTI-KPPQVVRVGSKKVAWINFKDICGIMHRPSE 110
Y Y+E+L R+ ++ NNP+LAG +R T+ +PPQV+R G+KK ++NF D+C MHR +
Sbjct 116 YIYDELLGRVFNILRENNPELAGDRRRTVMRPPQVLREGTKKTVFVNFMDLCKTMHRQPD 175
Query 111 HVLQFVLAELGT 122
HV+Q++LAELGT
Sbjct 176 HVMQYLLAELGT 187
> cel:K04G2.1 iftb-1; eIFTwoBeta (eIF2beta translation initiation
factor) family member (iftb-1); K03238 translation initiation
factor 2 subunit 2
Length=250
Score = 81.6 bits (200), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 38/72 (52%), Positives = 51/72 (70%), Gaps = 1/72 (1%)
Query 52 YSYEEMLKRIQELIVRNNPDLAGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPSE 110
Y+YEE L + +++ NPD AG K+ + IK P+V R GSKK A+ NF +IC +M R +
Sbjct 87 YTYEEALTLVYQVMKDKNPDFAGDKKKFAIKLPEVARAGSKKTAFSNFLEICRLMKRQDK 146
Query 111 HVLQFVLAELGT 122
HVLQF+LAELGT
Sbjct 147 HVLQFLLAELGT 158
> ath:AT3G07920 translation initiation factor; K03238 translation
initiation factor 2 subunit 2
Length=164
Score = 65.9 bits (159), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 33/73 (45%), Positives = 48/73 (65%), Gaps = 5/73 (6%)
Query 55 EEMLKRIQELIVRNNPDLAGSKRYTIK-PPQVVRV----GSKKVAWINFKDICGIMHRPS 109
+E+L+R+ ++ N+P+L G TI PPQV+R G+KK ++NF D C MHR
Sbjct 17 QEILRRVFNILRENSPELVGIWLLTIIWPPQVLREETAKGTKKTVFVNFMDYCKTMHRNP 76
Query 110 EHVLQFVLAELGT 122
+HV+ F+LAELGT
Sbjct 77 DHVMAFLLAELGT 89
> sce:YPL237W SUI3; Beta subunit of the translation initiation
factor eIF2, involved in the identification of the start codon;
proposed to be involved in mRNA binding; K03238 translation
initiation factor 2 subunit 2
Length=285
Score = 62.0 bits (149), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 35/92 (38%), Positives = 51/92 (55%), Gaps = 15/92 (16%)
Query 54 YEEMLKRIQELIVRNNPDLAGSK---RYTIKPPQVVRVGSKKVAWINFKDICGIMHRPSE 110
Y E+L R ++ NNP+LAG + ++ I PP +R G KK + N +DI +HR E
Sbjct 131 YSELLSRFFNILRTNNPELAGDRSGPKFRIPPPVCLRDG-KKTIFSNIQDIAEKLHRSPE 189
Query 111 HVLQFVLAELGTESPQSNRPPKHLTSVKGEKR 142
H++Q++ AELGT SV G+KR
Sbjct 190 HLIQYLFAELGTSG-----------SVDGQKR 210
> ath:AT5G01940 eukaryotic translation initiation factor 2B family
protein / eIF-2B family protein; K03238 translation initiation
factor 2 subunit 2
Length=231
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 45/72 (62%), Gaps = 2/72 (2%)
Query 52 YSYEEMLKRIQELIVRNNPDLAGSK-RYTIKPPQVVRVGSKKVAWINFKDICGIMHRPSE 110
Y Y+E+L + + + + +++ + R + PPQ++ G+ V +NF D+C MHR +
Sbjct 73 YGYKELLSMVFDRLREEDVEVSTERPRTVMMPPQLLAEGTITVC-LNFADLCRTMHRKPD 131
Query 111 HVLQFVLAELGT 122
HV++F+LA++ T
Sbjct 132 HVMKFLLAQMET 143
> xla:443810 rcc1-b, MGC115703, MGC132085, RanGEF, chc1, chc1-b,
rcc1, xrcc1; regulator of chromosome condensation 1; K11493
regulator of chromosome condensation
Length=420
Score = 32.3 bits (72), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 35/64 (54%), Gaps = 8/64 (12%)
Query 83 PQVVRVGSKKVAWINFKDI-CGIMHRPS----EHVLQFVLA---ELGTESPQSNRPPKHL 134
PQ + + +K ++F+D+ CG + HV F L+ +LGT++ QS P++L
Sbjct 228 PQCIHLKAKGSGRVHFQDVFCGAYFTFAVSQEGHVFGFGLSNYHQLGTKTTQSCYAPQNL 287
Query 135 TSVK 138
TS K
Sbjct 288 TSFK 291
> cel:Y71D11A.1 cdh-12; CaDHerin family member (cdh-12)
Length=2922
Score = 30.8 bits (68), Expect = 2.4, Method: Composition-based stats.
Identities = 15/41 (36%), Positives = 26/41 (63%), Gaps = 1/41 (2%)
Query 5 EHRAAQAFDFGERRKKKKEKKPEDQA-AEAASEAFVDGSGQ 44
+ +++Q R + KK K+PED+A ++ +E VDG+GQ
Sbjct 667 KKQSSQEVGGASRSEDKKTKEPEDEAESDTVTEKKVDGAGQ 707
> sce:YHR013C ARD1, NAA10; Ard1p (EC:2.3.1.88); K00670 peptide
alpha-N-acetyltransferase [EC:2.3.1.88]
Length=238
Score = 29.3 bits (64), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 14/48 (29%), Positives = 25/48 (52%), Gaps = 1/48 (2%)
Query 108 PSEHVLQFVLAELGTESPQSNRPPK-HLTSVKGEKRKRKRSQKTRLLR 154
P E ++ +VL ++ + Q N PP H+TS+ + R+ L+R
Sbjct 88 PGEKLVGYVLVKMNDDPDQQNEPPNGHITSLSVMRTYRRMGIAENLMR 135
> tgo:TGME49_114740 splicing factor 3B subunit 2, putative ; K12829
splicing factor 3B subunit 2
Length=743
Score = 29.3 bits (64), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 32/114 (28%), Positives = 46/114 (40%), Gaps = 17/114 (14%)
Query 29 QAAEAASEA-FVDGSGQLFVRGHVYSYEEMLKRIQELIVRNNPDLAGS-----KRYTIKP 82
QA A E F++G L G S L +R PD A S K YTI
Sbjct 511 QAGVAGGETPFLEGISSL---GGATSLSSGLDSPASFDLRRRPDGASSVASSAKPYTILT 567
Query 83 PQVVRVGSKKVAWINFKDICGIMHRPSEHVLQFVLAELGTESPQSNRPPKHLTS 136
PQ V VG ++ + G+ H + + + GT +P + P++L S
Sbjct 568 PQDVPVGQQQ------GQLFGVKH--TYKIPSTLPGNAGTATPSGHLTPRNLLS 613
> ath:AT5G61620 myb family transcription factor
Length=317
Score = 28.9 bits (63), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 28/58 (48%), Gaps = 2/58 (3%)
Query 7 RAAQAFDFGERRKKKKEKKPEDQAAEAASEAFVDGSGQLFVRGHVYSYEEMLKRIQEL 64
R A FD +K+KE+ +D + + + + G Q V+GH + E+ R Q L
Sbjct 165 RRASLFDISLEDQKEKERNSQDASTKTPPKQPITGIQQPVVQGHTQT--EISNRFQNL 220
Lambda K H
0.317 0.132 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4222647260
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40