bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1262_orf4
Length=324
Score E
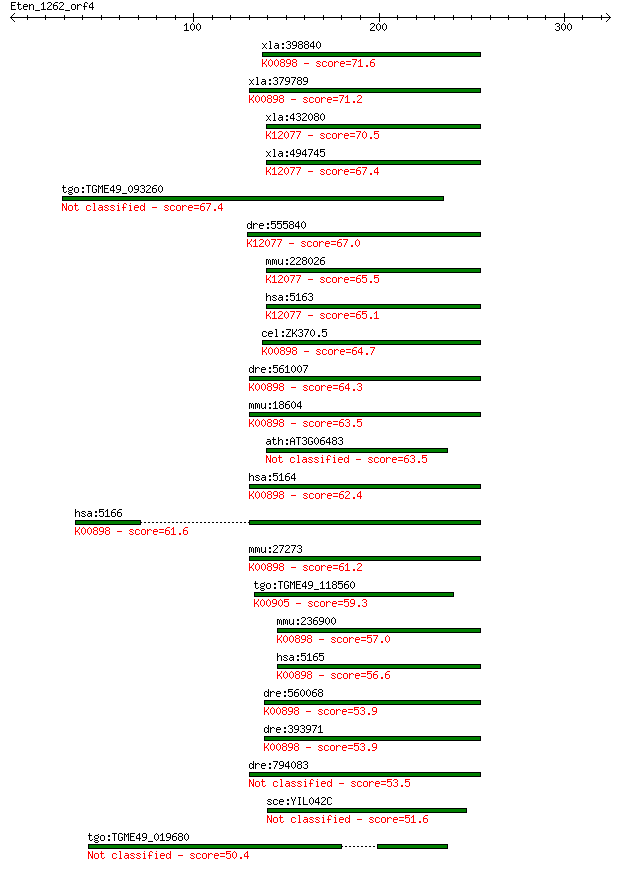
Sequences producing significant alignments: (Bits) Value
xla:398840 hypothetical protein MGC68579; K00898 pyruvate dehy... 71.6 4e-12
xla:379789 pdk4, 3j828, MGC132353, MGC52849; pyruvate dehydrog... 71.2 5e-12
xla:432080 pdk1, MGC81400; pyruvate dehydrogenase kinase, isoz... 70.5 9e-12
xla:494745 hypothetical LOC494745; K12077 pyruvate dehydrogena... 67.4 7e-11
tgo:TGME49_093260 pyruvate dehydrogenase (lipoamide) kinase, p... 67.4 7e-11
dre:555840 pyruvate dehydrogenase kinase 1-like; K12077 pyruva... 67.0 9e-11
mmu:228026 Pdk1, B830012B01, D530020C15Rik; pyruvate dehydroge... 65.5 3e-10
hsa:5163 PDK1; pyruvate dehydrogenase kinase, isozyme 1 (EC:2.... 65.1 4e-10
cel:ZK370.5 pdhk-2; Pyruvate DeHydogenase Kinase family member... 64.7 5e-10
dre:561007 si:rp71-57j15.4 (EC:2.7.11.2); K00898 pyruvate dehy... 64.3 6e-10
mmu:18604 Pdk2; pyruvate dehydrogenase kinase, isoenzyme 2 (EC... 63.5 1e-09
ath:AT3G06483 PDK; PDK (PYRUVATE DEHYDROGENASE KINASE); ATP bi... 63.5 1e-09
hsa:5164 PDK2, PDHK2; pyruvate dehydrogenase kinase, isozyme 2... 62.4 2e-09
hsa:5166 PDK4, FLJ40832; pyruvate dehydrogenase kinase, isozym... 61.6 4e-09
mmu:27273 Pdk4, AV005916; pyruvate dehydrogenase kinase, isoen... 61.2 5e-09
tgo:TGME49_118560 3-methyl-2-oxobutanoate dehydrogenase (lipoa... 59.3 2e-08
mmu:236900 Pdk3, 2610001M10Rik, AI035637, MGC6383; pyruvate de... 57.0 1e-07
hsa:5165 PDK3; pyruvate dehydrogenase kinase, isozyme 3 (EC:2.... 56.6 1e-07
dre:560068 pdk3a, pdk3; pyruvate dehydrogenase kinase, isozyme... 53.9 9e-07
dre:393971 pdk2, MGC56209, Pdk, zgc:56209; pyruvate dehydrogen... 53.9 9e-07
dre:794083 pyruvate dehydrogenase kinase 2-like 53.5 1e-06
sce:YIL042C PKP1; Mitochondrial protein kinase involved in neg... 51.6 4e-06
tgo:TGME49_019680 hypothetical protein 50.4 8e-06
> xla:398840 hypothetical protein MGC68579; K00898 pyruvate dehydrogenase
kinase [EC:2.7.11.2]
Length=404
Score = 71.6 bits (174), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 37/118 (31%), Positives = 66/118 (55%), Gaps = 7/118 (5%)
Query 137 KLPPVDIHLILAAESGVVVKVCDSGNGLCLTEQQLAWRFLYSTRTMPVDRQNKTRAGSAA 196
+LPP+ ++++L E V +K+ D+G G+ L + + + ++YST P+ + +R A
Sbjct 269 RLPPIKVNVVLGNED-VTIKISDNGGGVPLRKIERLFSYMYSTAPRPL--MDNSRNAPLA 325
Query 197 ALFSGWGVGLPLTRMYARALGGDAFMESRIGRGTRSYLYVPDISPTACTASPQTNSAA 254
G+G GLP++R+YAR GD + S G GT + +Y+ +S + P N +A
Sbjct 326 ----GFGYGLPISRLYARYFQGDLMLHSMEGFGTDAVIYLKALSSESVERLPVFNKSA 379
> xla:379789 pdk4, 3j828, MGC132353, MGC52849; pyruvate dehydrogenase
kinase, isozyme 4 (EC:2.7.11.2); K00898 pyruvate dehydrogenase
kinase [EC:2.7.11.2]
Length=404
Score = 71.2 bits (173), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 36/125 (28%), Positives = 70/125 (56%), Gaps = 7/125 (5%)
Query 130 QQQQQAIKLPPVDIHLILAAESGVVVKVCDSGNGLCLTEQQLAWRFLYSTRTMPVDRQNK 189
+ + + +LPP+ ++++L +E + +K+ D+G G+ L + + + ++YST P+ +
Sbjct 262 ENHETSPRLPPIKVNVVLGSED-LTIKISDNGGGVPLRKIERLFSYMYSTAPRPL--MDN 318
Query 190 TRAGSAAALFSGWGVGLPLTRMYARALGGDAFMESRIGRGTRSYLYVPDISPTACTASPQ 249
+R A G+G GLP++R+YAR GD + S G GT + +Y+ +S + P
Sbjct 319 SRNAPLA----GFGYGLPISRLYARYFQGDLMLHSMEGFGTDAVIYLKALSSESVERLPV 374
Query 250 TNSAA 254
N +A
Sbjct 375 FNKSA 379
> xla:432080 pdk1, MGC81400; pyruvate dehydrogenase kinase, isozyme
1 (EC:2.7.11.2); K12077 pyruvate dehydrogenase kinase
isoform 1 [EC:2.7.11.2]
Length=412
Score = 70.5 bits (171), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 38/116 (32%), Positives = 64/116 (55%), Gaps = 7/116 (6%)
Query 139 PPVDIHLILAAESGVVVKVCDSGNGLCLTEQQLAWRFLYSTRTMPVDRQNKTRAGSAAAL 198
PP+ +H++L +E + VK+ D G G+ L + + + ++YST +P R +RA A
Sbjct 275 PPIKVHVVLGSED-LTVKLSDRGGGVPLRKIERLFNYMYSTAPLP--RMETSRATPLA-- 329
Query 199 FSGWGVGLPLTRMYARALGGDAFMESRIGRGTRSYLYVPDISPTACTASPQTNSAA 254
G+G GLP++R+YA+ GD + S G GT + +Y +S + P N +A
Sbjct 330 --GFGYGLPISRLYAKYFQGDLKLYSLEGYGTDAVIYFKALSTESVERLPVYNKSA 383
> xla:494745 hypothetical LOC494745; K12077 pyruvate dehydrogenase
kinase isoform 1 [EC:2.7.11.2]
Length=412
Score = 67.4 bits (163), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 38/116 (32%), Positives = 63/116 (54%), Gaps = 7/116 (6%)
Query 139 PPVDIHLILAAESGVVVKVCDSGNGLCLTEQQLAWRFLYSTRTMPVDRQNKTRAGSAAAL 198
PP+ +H+ L +E + VK+ D G G+ L + + + ++YST +P R +RA A
Sbjct 275 PPIKVHVALGSED-LSVKLSDRGGGVPLRKIERLFNYMYSTAPLP--RMETSRATPLA-- 329
Query 199 FSGWGVGLPLTRMYARALGGDAFMESRIGRGTRSYLYVPDISPTACTASPQTNSAA 254
G+G GLP++R+YA+ GD + S G GT + +Y +S + P N +A
Sbjct 330 --GFGYGLPISRLYAKYFQGDLKLYSLEGYGTDAVIYFKALSTESVERLPVYNKSA 383
> tgo:TGME49_093260 pyruvate dehydrogenase (lipoamide) kinase,
putative (EC:2.7.11.2)
Length=1105
Score = 67.4 bits (163), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 70/292 (23%), Positives = 116/292 (39%), Gaps = 96/292 (32%)
Query 29 SSRRHAADNRVVVRCAPAYVYSGCFELIKNAIEATVQRQQKQQKQQQSQQQQQEQQGAVE 88
S R V C +Y YSG FEL+KNA+ A+V +++ + + + E++ +E
Sbjct 758 SPRETGLPGTVRFPCVSSYAYSGVFELVKNAMRASVDSWTEREALEARSEVELEEERILE 817
Query 89 EEQ------------------------------------------QQEWDALPVRQSRCI 106
E+ + W L V +SR +
Sbjct 818 TERAIYTWRTSRERGSGIREEGGTTWRSTSSSLSSASLAAGNAVLRGPWSLLTVGRSRRL 877
Query 107 YSLPALYGAPSQEQQQLQLPQQQQQ----QQQAIKL-----PPVDIHLILAAESGVVVKV 157
LP +QL+ + Q+ + + + L V ++L++A S ++++V
Sbjct 878 RKLPG---------RQLESCEAQRDGCGGRTRVVVLEEDAPSSVKVNLVVAFGS-LLIQV 927
Query 158 CDSGNGLCLTEQQLAWRFLYSTR------TMPV-------------------------DR 186
D+G GL E W F YST PV D+
Sbjct 928 EDTGRGLAEDELAKIWSFAYSTAHGAGEAKGPVSTGMKHSGNQRKDEGGEQGDKGEKDDK 987
Query 187 QNK----TRAGSAAALFSGWGVGLPLTRMYARALGGDAFMESRIGRGTRSYL 234
Q K + A + +G GVGLP++R++A++LGGD F+ES G GT +Y+
Sbjct 988 QEKGGKEDDSDEAVPVLAGCGVGLPISRVHAQSLGGDIFIESAKGVGTCAYM 1039
> dre:555840 pyruvate dehydrogenase kinase 1-like; K12077 pyruvate
dehydrogenase kinase isoform 1 [EC:2.7.11.2]
Length=405
Score = 67.0 bits (162), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 40/126 (31%), Positives = 67/126 (53%), Gaps = 7/126 (5%)
Query 129 QQQQQQAIKLPPVDIHLILAAESGVVVKVCDSGNGLCLTEQQLAWRFLYSTRTMPVDRQN 188
+ + A++ PPV + ++L E + VKV D G G+ L +++ F Y+ T P + +
Sbjct 258 MELYEDAMEYPPVHVQIVLGHED-LTVKVSDRGGGVPL--RKIDRLFTYTYSTAPRPQMD 314
Query 189 KTRAGSAAALFSGWGVGLPLTRMYARALGGDAFMESRIGRGTRSYLYVPDISPTACTASP 248
+RA A G+G GLP++R+YAR GD + S G GT + +Y+ +S + P
Sbjct 315 TSRATPLA----GYGYGLPISRLYARYFQGDLKLYSMEGFGTDAVIYIRALSTDSIERLP 370
Query 249 QTNSAA 254
N +A
Sbjct 371 VYNKSA 376
> mmu:228026 Pdk1, B830012B01, D530020C15Rik; pyruvate dehydrogenase
kinase, isoenzyme 1 (EC:2.7.11.2); K12077 pyruvate dehydrogenase
kinase isoform 1 [EC:2.7.11.2]
Length=434
Score = 65.5 bits (158), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 39/116 (33%), Positives = 61/116 (52%), Gaps = 7/116 (6%)
Query 139 PPVDIHLILAAESGVVVKVCDSGNGLCLTEQQLAWRFLYSTRTMPVDRQNKTRAGSAAAL 198
PP+ +H+ L E + VK+ D G G+ L + + ++YST P R +RA A
Sbjct 297 PPIQVHVTLGEED-LTVKMSDRGGGVPLRKIDRLFNYMYSTAPRP--RVETSRAVPLA-- 351
Query 199 FSGWGVGLPLTRMYARALGGDAFMESRIGRGTRSYLYVPDISPTACTASPQTNSAA 254
G+G GLP++R+YA+ GD + S G GT + +Y+ +S + P N AA
Sbjct 352 --GFGYGLPISRLYAQYFQGDLKLYSLEGYGTDAVIYIKALSTESVERLPVYNKAA 405
> hsa:5163 PDK1; pyruvate dehydrogenase kinase, isozyme 1 (EC:2.7.11.2);
K12077 pyruvate dehydrogenase kinase isoform 1 [EC:2.7.11.2]
Length=436
Score = 65.1 bits (157), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 39/116 (33%), Positives = 61/116 (52%), Gaps = 7/116 (6%)
Query 139 PPVDIHLILAAESGVVVKVCDSGNGLCLTEQQLAWRFLYSTRTMPVDRQNKTRAGSAAAL 198
PP+ +H+ L E + VK+ D G G+ L + + ++YST P R +RA A
Sbjct 299 PPIQVHVTLGNED-LTVKMSDRGGGVPLRKIDRLFNYMYSTAPRP--RVETSRAVPLA-- 353
Query 199 FSGWGVGLPLTRMYARALGGDAFMESRIGRGTRSYLYVPDISPTACTASPQTNSAA 254
G+G GLP++R+YA+ GD + S G GT + +Y+ +S + P N AA
Sbjct 354 --GFGYGLPISRLYAQYFQGDLKLYSLEGYGTDAVIYIKALSTDSIERLPVYNKAA 407
> cel:ZK370.5 pdhk-2; Pyruvate DeHydogenase Kinase family member
(pdhk-2); K00898 pyruvate dehydrogenase kinase [EC:2.7.11.2]
Length=401
Score = 64.7 bits (156), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 37/118 (31%), Positives = 64/118 (54%), Gaps = 8/118 (6%)
Query 137 KLPPVDIHLILAAESGVVVKVCDSGNGLCLTEQQLAWRFLYSTRTMPVDRQNKTRAGSAA 196
LP + ++++ E + +K+CD G G+ T + + ++YST P R G+ A
Sbjct 265 DLPDIKVYVVKGQED-LSIKICDRGGGVSRTILERLYNYMYSTAPPP------PRDGTQA 317
Query 197 ALFSGWGVGLPLTRMYARALGGDAFMESRIGRGTRSYLYVPDISPTACTASPQTNSAA 254
L +G+G GLPL+R+YAR GD F+ S G GT + +Y+ + A P ++++
Sbjct 318 PL-AGYGYGLPLSRLYARYFLGDLFLVSMEGHGTDACIYLKAVPVEASEVLPIYSTSS 374
> dre:561007 si:rp71-57j15.4 (EC:2.7.11.2); K00898 pyruvate dehydrogenase
kinase [EC:2.7.11.2]
Length=409
Score = 64.3 bits (155), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 35/125 (28%), Positives = 66/125 (52%), Gaps = 7/125 (5%)
Query 130 QQQQQAIKLPPVDIHLILAAESGVVVKVCDSGNGLCLTEQQLAWRFLYSTRTMPVDRQNK 189
+ + ++ LPP+ + + L E + +K+ D G+G+ L + + + ++YST PV +
Sbjct 262 ETHETSLHLPPIKVRVSLGTED-LTIKMSDRGSGVPLRKIERLFSYMYSTAPSPVAEDTR 320
Query 190 TRAGSAAALFSGWGVGLPLTRMYARALGGDAFMESRIGRGTRSYLYVPDISPTACTASPQ 249
A +G+G GLP++R+YA+ GD + S G GT + +Y+ +S + P
Sbjct 321 N------APLAGFGYGLPISRLYAKYFQGDLQLYSMEGYGTSAVIYLKALSTESIERLPV 374
Query 250 TNSAA 254
N +A
Sbjct 375 FNKSA 379
> mmu:18604 Pdk2; pyruvate dehydrogenase kinase, isoenzyme 2 (EC:2.7.11.2);
K00898 pyruvate dehydrogenase kinase [EC:2.7.11.2]
Length=407
Score = 63.5 bits (153), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 34/125 (27%), Positives = 65/125 (52%), Gaps = 8/125 (6%)
Query 130 QQQQQAIKLPPVDIHLILAAESGVVVKVCDSGNGLCLTEQQLAWRFLYSTRTMPVDRQNK 189
+ + ++ LPP+ I ++ E + +K+ D G G+ L + + + ++YST P
Sbjct 262 ESHESSLTLPPIKI-MVALGEEDLSIKMSDRGGGVPLRKIERLFSYMYSTAPTP------ 314
Query 190 TRAGSAAALFSGWGVGLPLTRMYARALGGDAFMESRIGRGTRSYLYVPDISPTACTASPQ 249
+ G+ +G+G GLP++R+YA+ GD + S G GT + +Y+ +S + P
Sbjct 315 -QPGTGGTPLAGFGYGLPISRLYAKYFQGDLQLFSMEGFGTDAVIYLKALSTDSVERLPV 373
Query 250 TNSAA 254
N +A
Sbjct 374 YNKSA 378
> ath:AT3G06483 PDK; PDK (PYRUVATE DEHYDROGENASE KINASE); ATP
binding / histidine phosphotransfer kinase/ pyruvate dehydrogenase
(acetyl-transferring) kinase
Length=366
Score = 63.5 bits (153), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 32/98 (32%), Positives = 51/98 (52%), Gaps = 1/98 (1%)
Query 139 PPVDIHLILAAESGVVVKVCDSGNGLCLTEQQLAWRFLYSTRTMPVDRQNKTRAGSAAAL 198
PP+ I ++ V +KV D G G+ + + +LYST P++
Sbjct 259 PPIRI-IVADGIEDVTIKVSDEGGGIARSGLPRIFTYLYSTARNPLEEDVDLGIADVPVT 317
Query 199 FSGWGVGLPLTRMYARALGGDAFMESRIGRGTRSYLYV 236
+G+G GLP++R+YAR GGD + S G GT +YL++
Sbjct 318 MAGYGYGLPISRLYARYFGGDLQIISMEGYGTDAYLHL 355
> hsa:5164 PDK2, PDHK2; pyruvate dehydrogenase kinase, isozyme
2 (EC:2.7.11.2); K00898 pyruvate dehydrogenase kinase [EC:2.7.11.2]
Length=343
Score = 62.4 bits (150), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 33/125 (26%), Positives = 65/125 (52%), Gaps = 8/125 (6%)
Query 130 QQQQQAIKLPPVDIHLILAAESGVVVKVCDSGNGLCLTEQQLAWRFLYSTRTMPVDRQNK 189
+ + ++ LPP+ + ++ E + +K+ D G G+ L + + + ++YST P
Sbjct 198 ESHESSLILPPIKV-MVALGEEDLSIKMSDRGGGVPLRKIERLFSYMYSTAPTP------ 250
Query 190 TRAGSAAALFSGWGVGLPLTRMYARALGGDAFMESRIGRGTRSYLYVPDISPTACTASPQ 249
+ G+ +G+G GLP++R+YA+ GD + S G GT + +Y+ +S + P
Sbjct 251 -QPGTGGTPLAGFGYGLPISRLYAKYFQGDLQLFSMEGFGTDAVIYLKALSTDSVERLPV 309
Query 250 TNSAA 254
N +A
Sbjct 310 YNKSA 314
> hsa:5166 PDK4, FLJ40832; pyruvate dehydrogenase kinase, isozyme
4 (EC:2.7.11.2); K00898 pyruvate dehydrogenase kinase [EC:2.7.11.2]
Length=411
Score = 61.6 bits (148), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 36/125 (28%), Positives = 62/125 (49%), Gaps = 7/125 (5%)
Query 130 QQQQQAIKLPPVDIHLILAAESGVVVKVCDSGNGLCLTEQQLAWRFLYSTRTMPVDRQNK 189
+ Q+ L P+++ ++L E + +K+ D G G+ L + + YST PV
Sbjct 265 EHQENQPSLTPIEVIVVLGKED-LTIKISDRGGGVPLRIIDRLFSYTYSTAPTPV----- 318
Query 190 TRAGSAAALFSGWGVGLPLTRMYARALGGDAFMESRIGRGTRSYLYVPDISPTACTASPQ 249
S A +G+G GLP++R+YA+ GD + S G GT + +Y+ +S + P
Sbjct 319 -MDNSRNAPLAGFGYGLPISRLYAKYFQGDLNLYSLSGYGTDAIIYLKALSSESIEKLPV 377
Query 250 TNSAA 254
N +A
Sbjct 378 FNKSA 382
Score = 32.0 bits (71), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 13/35 (37%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 36 DNRVVVRCAPAYVYSGCFELIKNAIEATVQRQQKQ 70
D + + P++++ FEL KNA+ ATV+ Q+ Q
Sbjct 236 DQPIHIVYVPSHLHHMLFELFKNAMRATVEHQENQ 270
> mmu:27273 Pdk4, AV005916; pyruvate dehydrogenase kinase, isoenzyme
4 (EC:2.7.11.2); K00898 pyruvate dehydrogenase kinase
[EC:2.7.11.2]
Length=412
Score = 61.2 bits (147), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 37/125 (29%), Positives = 61/125 (48%), Gaps = 7/125 (5%)
Query 130 QQQQQAIKLPPVDIHLILAAESGVVVKVCDSGNGLCLTEQQLAWRFLYSTRTMPVDRQNK 189
+ Q+ L PV+ ++L E + +K+ D G G+ L + + YST PV
Sbjct 265 EHQENRPSLTPVEATVVLGKED-LTIKISDRGGGVPLRITDRLFSYTYSTAPTPV----- 318
Query 190 TRAGSAAALFSGWGVGLPLTRMYARALGGDAFMESRIGRGTRSYLYVPDISPTACTASPQ 249
S A +G+G GLP++R+YA+ GD + S G GT + +Y+ +S + P
Sbjct 319 -MDNSRNAPLAGFGYGLPISRLYAKYFQGDLNLYSMSGYGTDAIIYLKALSSESVEKLPV 377
Query 250 TNSAA 254
N +A
Sbjct 378 FNKSA 382
> tgo:TGME49_118560 3-methyl-2-oxobutanoate dehydrogenase (lipoamide)
kinase, putative (EC:2.7.11.4); K00905 [3-methyl-2-oxobutanoate
dehydrogenase (acetyl-transferring)] kinase [EC:2.7.11.4]
Length=432
Score = 59.3 bits (142), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 35/111 (31%), Positives = 53/111 (47%), Gaps = 5/111 (4%)
Query 133 QQAIKLPPVDIHLILAAESGVVVKVCDSGNGLCLTEQQLAWRFLYSTRTMPVDRQNKTRA 192
Q+ LP V + + + VV+K+ D G G+ + Q W F YST + + +
Sbjct 300 QEDEDLPEVKVE-VYKGKREVVIKISDKGGGVPPPKLQDIWSFGYSTVGDSNTKMQENSS 358
Query 193 GSAAALF----SGWGVGLPLTRMYARALGGDAFMESRIGRGTRSYLYVPDI 239
G +G+G GLPL R +AR GGD ++S G GT Y+ + I
Sbjct 359 GLGENFIRSDMAGYGFGLPLARAFARYFGGDIHVQSHFGIGTDVYITLNHI 409
> mmu:236900 Pdk3, 2610001M10Rik, AI035637, MGC6383; pyruvate
dehydrogenase kinase, isoenzyme 3 (EC:2.7.11.2); K00898 pyruvate
dehydrogenase kinase [EC:2.7.11.2]
Length=415
Score = 57.0 bits (136), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 34/110 (30%), Positives = 55/110 (50%), Gaps = 6/110 (5%)
Query 145 LILAAESGVVVKVCDSGNGLCLTEQQLAWRFLYSTRTMPVDRQNKTRAGSAAALFSGWGV 204
L+ + + +K+ D G G+ L + + ++YST P TRA A G+G
Sbjct 273 LVTLGKEDLSIKISDLGGGVPLRKIDRLFNYMYSTAPRP--SLEPTRAAPLA----GFGY 326
Query 205 GLPLTRMYARALGGDAFMESRIGRGTRSYLYVPDISPTACTASPQTNSAA 254
GLP++R+YAR GD + S G GT + +Y+ +S + P N +A
Sbjct 327 GLPISRLYARYFQGDLKLYSMEGVGTDAVIYLKALSSESFERLPVFNKSA 376
> hsa:5165 PDK3; pyruvate dehydrogenase kinase, isozyme 3 (EC:2.7.11.2);
K00898 pyruvate dehydrogenase kinase [EC:2.7.11.2]
Length=415
Score = 56.6 bits (135), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 34/110 (30%), Positives = 55/110 (50%), Gaps = 6/110 (5%)
Query 145 LILAAESGVVVKVCDSGNGLCLTEQQLAWRFLYSTRTMPVDRQNKTRAGSAAALFSGWGV 204
L+ + + +K+ D G G+ L + + ++YST P TRA A G+G
Sbjct 273 LVTLGKEDLSIKISDLGGGVPLRKIDRLFNYMYSTAPRP--SLEPTRAAPLA----GFGY 326
Query 205 GLPLTRMYARALGGDAFMESRIGRGTRSYLYVPDISPTACTASPQTNSAA 254
GLP++R+YAR GD + S G GT + +Y+ +S + P N +A
Sbjct 327 GLPISRLYARYFQGDLKLYSMEGVGTDAVIYLKALSSESFERLPVFNKSA 376
> dre:560068 pdk3a, pdk3; pyruvate dehydrogenase kinase, isozyme
3a; K00898 pyruvate dehydrogenase kinase [EC:2.7.11.2]
Length=404
Score = 53.9 bits (128), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 35/117 (29%), Positives = 55/117 (47%), Gaps = 8/117 (6%)
Query 138 LPPVDIHLILAAESGVVVKVCDSGNGLCLTEQQLAWRFLYSTRTMPVDRQNKTRAGSAAA 197
LP + + L E + VK+ D G G+ L + + + YST P S
Sbjct 266 LPLIKAKVTLGIED-LSVKISDRGGGVALRKIDRLFNYTYSTAPTP-------SLDSKRV 317
Query 198 LFSGWGVGLPLTRMYARALGGDAFMESRIGRGTRSYLYVPDISPTACTASPQTNSAA 254
+G+G GLP++R+YAR GD + S G GT + +Y+ +S + P N +A
Sbjct 318 PLAGFGHGLPISRLYARYFQGDLKLYSMEGVGTDAVIYLKALSSESFERLPVFNKSA 374
> dre:393971 pdk2, MGC56209, Pdk, zgc:56209; pyruvate dehydrogenase
kinase, isoenzyme 2 (EC:2.7.11.2); K00898 pyruvate dehydrogenase
kinase [EC:2.7.11.2]
Length=409
Score = 53.9 bits (128), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 34/119 (28%), Positives = 56/119 (47%), Gaps = 11/119 (9%)
Query 138 LPPVDIHLILAAESGVVVKVCDSGNGLCLTEQQLAWRFLYSTRTMPV--DRQNKTRAGSA 195
LPP+ + + L E + +K+ D G G+ + + + ++YST P D Q AG
Sbjct 270 LPPIKVMVALGGED-LSIKISDRGGGVPFRKIERLFSYMYSTAPRPTIGDHQRTPMAG-- 326
Query 196 AALFSGWGVGLPLTRMYARALGGDAFMESRIGRGTRSYLYVPDISPTACTASPQTNSAA 254
+G GLP++R+YAR GD + G GT + + + +S + P N A
Sbjct 327 ------FGYGLPISRLYARYFQGDLQLYPMEGYGTDAVIQLKALSTDSVEKLPVFNKTA 379
> dre:794083 pyruvate dehydrogenase kinase 2-like
Length=409
Score = 53.5 bits (127), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 33/125 (26%), Positives = 61/125 (48%), Gaps = 7/125 (5%)
Query 130 QQQQQAIKLPPVDIHLILAAESGVVVKVCDSGNGLCLTEQQLAWRFLYSTRTMPVDRQNK 189
+ ++ LP + + + + E + +K+ D G G+ + + + ++YST P K
Sbjct 262 ENHKEGSNLPAIQVMVAVGGED-LTIKMSDRGGGVPFRKMENLFSYMYSTAPTP-QMDEK 319
Query 190 TRAGSAAALFSGWGVGLPLTRMYARALGGDAFMESRIGRGTRSYLYVPDISPTACTASPQ 249
RA A G+G GLP++R+YA+ GD + S G GT + +++ +S + P
Sbjct 320 QRAPLA-----GFGYGLPISRLYAKYFQGDLQLYSMEGHGTDAVIHLKALSTDSVERLPV 374
Query 250 TNSAA 254
N A
Sbjct 375 FNKTA 379
> sce:YIL042C PKP1; Mitochondrial protein kinase involved in negative
regulation of pyruvate dehydrogenase complex activity
by phosphorylating the ser-133 residue of the Pda1p subunit;
acts in concert with kinase Pkp2p and phosphatases Ptc5p
and Ptc6p (EC:2.7.11.2)
Length=394
Score = 51.6 bits (122), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 30/108 (27%), Positives = 53/108 (49%), Gaps = 1/108 (0%)
Query 140 PVDIHLILAAESGVVVKVCDSGNGLCLTEQQLAWRFLYSTRTM-PVDRQNKTRAGSAAAL 198
P++I+L+ + + +++ D G G+ + L + + YST T D ++ G
Sbjct 285 PIEINLLKPDDDELYLRIRDHGGGITPEVEALMFNYSYSTHTQQSADSESTDLPGEQINN 344
Query 199 FSGWGVGLPLTRMYARALGGDAFMESRIGRGTRSYLYVPDISPTACTA 246
SG G GLP+ + Y GG ++S +G GT Y+ + S TA +
Sbjct 345 VSGMGFGLPMCKTYLELFGGKIDVQSLLGWGTDVYIKLKGPSKTALLS 392
> tgo:TGME49_019680 hypothetical protein
Length=808
Score = 50.4 bits (119), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 42/153 (27%), Positives = 68/153 (44%), Gaps = 34/153 (22%)
Query 43 CAPAYVYSGCFELIKNAIEATVQRQQKQQKQQQSQQQQQEQQGAVEEEQQQEWDALPVRQ 102
C P Y+Y FEL KNA+ ATV+R ++S+ A +EE + + VR
Sbjct 458 CVPQYLYYILFELFKNAMRATVERFGADSSSRRSRSS------AFDEEAGKSFSG--VRT 509
Query 103 SRC----IYSLP-----------ALYGAPSQEQQQLQLPQQQQQQQQAIKLPPVDIHLIL 147
SR +SL YG P Q + +L ++LPP I L++
Sbjct 510 SRGSVGNAFSLKREEDYEYDSSFLFYGRPPQYETKL--------ADSDMQLPP--IQLVV 559
Query 148 AAESGVV-VKVCDSGNGLCLTEQQLAWRFLYST 179
+ ++ V+ +K+ D G G+ W ++Y+T
Sbjct 560 SGDNRVIAIKMSDQGGGVAQESIDKIWSYMYTT 592
Score = 37.7 bits (86), Expect = 0.053, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 25/38 (65%), Gaps = 0/38 (0%)
Query 199 FSGWGVGLPLTRMYARALGGDAFMESRIGRGTRSYLYV 236
+G+G GLPL+R+YA LGG + S G+ +YLY+
Sbjct 731 LAGFGCGLPLSRLYASYLGGRLEILSLPFHGSDAYLYL 768
Lambda K H
0.316 0.125 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 13394984740
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40