bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1252_orf2
Length=205
Score E
Sequences producing significant alignments: (Bits) Value
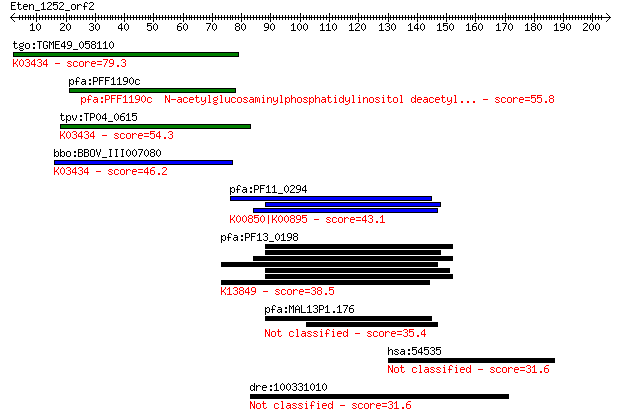
tgo:TGME49_058110 N-acetylglucosaminylphosphatidylinositol dea... 79.3 7e-15
pfa:PFF1190c N-acetylglucosaminylphosphatidylinositol deacetyl... 55.8 1e-07
tpv:TP04_0615 N-acetylglucosaminyl-phosphatidylinositol de-N-a... 54.3 3e-07
bbo:BBOV_III007080 17.m07624; N-acetylglucosaminyl-phosphatidy... 46.2 7e-05
pfa:PF11_0294 ATP-dependent phosphofructokinase, putative; K00... 43.1 7e-04
pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog A... 38.5 0.017
pfa:MAL13P1.176 PfRh2b; reticulocyte binding protein 2, homolog B 35.4 0.16
hsa:54535 CCHCR1, C6orf18, HCR, MGC126371, MGC126372, SBP; coi... 31.6 1.9
dre:100331010 upstream stimulatory factor 1-like 31.6 2.2
> tgo:TGME49_058110 N-acetylglucosaminylphosphatidylinositol deacetylase,
putative (EC:3.5.1.89); K03434 N-acetylglucosaminylphosphatidylinositol
deacetylase [EC:3.5.1.89]
Length=304
Score = 79.3 bits (194), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 36/77 (46%), Positives = 48/77 (62%), Gaps = 1/77 (1%)
Query 2 ELAAAAAAFCVPKENFVCLDEKGMKDGWTKWDSKAVCAAVKSFVEKRKQIQVIFSFDELG 61
E AA F V N + LD++ ++DGW W + V V+ F+EK +I IF+FDE G
Sbjct 182 EFLNAARLFGVENGNALVLDDEALQDGWGLWSPERVADVVEEFIEK-NEISTIFTFDERG 240
Query 62 VSRHPNHCSVYEGLRLY 78
VSRHPNH SV+ G+R Y
Sbjct 241 VSRHPNHISVFRGVRKY 257
> pfa:PFF1190c N-acetylglucosaminylphosphatidylinositol deacetylase,
putative (EC:3.5.1.89); K01463 [EC:3.5.1.-]
Length=257
Score = 55.8 bits (133), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 22/57 (38%), Positives = 35/57 (61%), Gaps = 1/57 (1%)
Query 21 DEKGMKDGWTKWDSKAVCAAVKSFVEKRKQIQVIFSFDELGVSRHPNHCSVYEGLRL 77
++ ++DGW WD K + +K + + I+ IF+FD GVS HPNH S Y+ +R+
Sbjct 120 NDNKIQDGWLYWDEKYIFKLIKDYCIQY-DIKTIFTFDNYGVSGHPNHISAYKSIRM 175
> tpv:TP04_0615 N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase
(EC:3.5.1.89); K03434 N-acetylglucosaminylphosphatidylinositol
deacetylase [EC:3.5.1.89]
Length=273
Score = 54.3 bits (129), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 39/65 (60%), Gaps = 1/65 (1%)
Query 18 VCLDEKGMKDGWTKWDSKAVCAAVKSFVEKRKQIQVIFSFDELGVSRHPNHCSVYEGLRL 77
+++ ++DG TKWD + VK F+ ++V+F+FD GVS HPNH S YE +L
Sbjct 136 TVINDPLLQDGDTKWDPEHALPHVKKFINDH-SLRVLFTFDGHGVSGHPNHISTYETAKL 194
Query 78 YNAEI 82
+ E+
Sbjct 195 ASKEL 199
> bbo:BBOV_III007080 17.m07624; N-acetylglucosaminyl-phosphatidylinositol
de-n-acetylase; K03434 N-acetylglucosaminylphosphatidylinositol
deacetylase [EC:3.5.1.89]
Length=253
Score = 46.2 bits (108), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 39/61 (63%), Gaps = 1/61 (1%)
Query 16 NFVCLDEKGMKDGWTKWDSKAVCAAVKSFVEKRKQIQVIFSFDELGVSRHPNHCSVYEGL 75
N + ++ +KDG W+ +AV V+ F+ + +++F+FDE GVS HPNH SV++ +
Sbjct 112 NCIIDNDPSVKDGPDDWNIEAVSHRVEDFI-RSVNAKMVFTFDEHGVSGHPNHKSVHKAV 170
Query 76 R 76
+
Sbjct 171 K 171
> pfa:PF11_0294 ATP-dependent phosphofructokinase, putative; K00850
6-phosphofructokinase [EC:2.7.1.11]; K00895 pyrophosphate--fructose-6-phosphate
1-phosphotransferase [EC:2.7.1.90]
Length=1570
Score = 43.1 bits (100), Expect = 7e-04, Method: Composition-based stats.
Identities = 21/69 (30%), Positives = 37/69 (53%), Gaps = 0/69 (0%)
Query 76 RLYNAEILETCKEQQKQHQQQRKQEHKQQQQQQLQLRKDEAQADSEAGTQRKGRQQEAED 135
++ + E+ E KE+ K Q K E ++QQ +Q + E Q + + K +Q E D
Sbjct 258 KIKSQELKEKNKEETKCKQDGEKNEKEEQQNEQDDEKNKEQQKCKQDDEKNKEQQNEQRD 317
Query 136 EQQQREQQQ 144
++Q +EQQ+
Sbjct 318 DEQNKEQQK 326
Score = 41.2 bits (95), Expect = 0.003, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 36/60 (60%), Gaps = 0/60 (0%)
Query 88 EQQKQHQQQRKQEHKQQQQQQLQLRKDEAQADSEAGTQRKGRQQEAEDEQQQREQQQMQD 147
+++ + QQ+ KQ+ ++ ++QQ + R DE + + Q + +E ++EQ+ EQ M+D
Sbjct 292 DEKNKEQQKCKQDDEKNKEQQNEQRDDEQNKEQQKCKQDDEQNKEQQNEQRDDEQINMED 351
Score = 38.5 bits (88), Expect = 0.015, Method: Composition-based stats.
Identities = 17/63 (26%), Positives = 37/63 (58%), Gaps = 0/63 (0%)
Query 84 ETCKEQQKQHQQQRKQEHKQQQQQQLQLRKDEAQADSEAGTQRKGRQQEAEDEQQQREQQ 143
E +++++Q++Q ++ +QQ+ +Q + E Q + Q K +Q+ +D++Q +EQQ
Sbjct 279 EKNEKEEQQNEQDDEKNKEQQKCKQDDEKNKEQQNEQRDDEQNKEQQKCKQDDEQNKEQQ 338
Query 144 QMQ 146
Q
Sbjct 339 NEQ 341
> pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog
A; K13849 reticulocyte-binding protein
Length=3130
Score = 38.5 bits (88), Expect = 0.017, Method: Composition-based stats.
Identities = 17/64 (26%), Positives = 42/64 (65%), Gaps = 0/64 (0%)
Query 88 EQQKQHQQQRKQEHKQQQQQQLQLRKDEAQADSEAGTQRKGRQQEAEDEQQQREQQQMQD 147
++Q+Q + Q+++ K+Q+Q++LQ ++ + + E + K Q + E+E +++EQ+++Q
Sbjct 2776 KRQEQERLQKEEALKRQEQERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQK 2835
Query 148 MQLL 151
+ L
Sbjct 2836 EEAL 2839
Score = 38.5 bits (88), Expect = 0.018, Method: Composition-based stats.
Identities = 20/60 (33%), Positives = 42/60 (70%), Gaps = 4/60 (6%)
Query 88 EQQKQHQQQRKQEHKQQQQQQLQLRKDEAQADSEAGTQRKGRQQEAEDEQQQREQQQMQD 147
E++KQ Q Q+++E K+Q+Q++LQ K+EA E +R +++E + ++Q+R +++ Q+
Sbjct 2762 EREKQEQLQKEEELKRQEQERLQ--KEEALKRQE--QERLQKEEELKRQEQERLEREKQE 2817
Score = 36.6 bits (83), Expect = 0.065, Method: Composition-based stats.
Identities = 22/69 (31%), Positives = 43/69 (62%), Gaps = 2/69 (2%)
Query 84 ETCKEQQKQHQQQRKQEHKQQQQQQLQLRKDEA-QADSEAGTQRKGRQQEAEDEQQQREQ 142
E ++Q+Q + Q+++E K+Q+Q++L+ K E Q + E Q + R Q+ E ++Q EQ
Sbjct 2786 EEALKRQEQERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQ-EQ 2844
Query 143 QQMQDMQLL 151
+++Q + L
Sbjct 2845 ERLQKEEEL 2853
Score = 36.2 bits (82), Expect = 0.076, Method: Composition-based stats.
Identities = 18/76 (23%), Positives = 48/76 (63%), Gaps = 2/76 (2%)
Query 73 EGLRLYNAEILETCKEQQKQHQQQRKQEHKQQQQQQLQLRKDEAQ--ADSEAGTQRKGRQ 130
E L+ E L+ +E ++Q Q++ ++E ++Q Q++ +L++ E + EA +++ +
Sbjct 2787 EALKRQEQERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQER 2846
Query 131 QEAEDEQQQREQQQMQ 146
+ E+E +++EQ++++
Sbjct 2847 LQKEEELKRQEQERLE 2862
Score = 36.2 bits (82), Expect = 0.085, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 43/63 (68%), Gaps = 6/63 (9%)
Query 88 EQQKQHQQQRKQEHKQQQQQQLQLRKDEAQADSEAGTQRKGRQQEAEDEQQQREQQQMQD 147
E++KQ Q Q+++E K+Q+Q++LQ K+EA E ++ Q+E E ++Q++E+ + +
Sbjct 2812 EREKQEQLQKEEELKRQEQERLQ--KEEALKRQE----QERLQKEEELKRQEQERLERKK 2865
Query 148 MQL 150
++L
Sbjct 2866 IEL 2868
Score = 36.2 bits (82), Expect = 0.090, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 42/65 (64%), Gaps = 2/65 (3%)
Query 88 EQQKQHQQQRKQEHKQQQQQQLQLRKDEA-QADSEAGTQRKGRQQEAEDEQQQREQQQMQ 146
++Q+Q + Q+++E K+Q+Q++L+ K E Q + E Q + R Q+ E ++Q EQ+++Q
Sbjct 2740 KRQEQERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQ-EQERLQ 2798
Query 147 DMQLL 151
+ L
Sbjct 2799 KEEEL 2803
Score = 30.8 bits (68), Expect = 3.1, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 41/74 (55%), Gaps = 3/74 (4%)
Query 73 EGLRLYNAEILETCKEQQKQHQQQ-RKQEHKQQQQQQLQLRKDEA--QADSEAGTQRKGR 129
E L+ E LE K++Q Q +++ ++QE ++ Q+++ R+++ Q + E Q + R
Sbjct 2801 EELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELKRQEQER 2860
Query 130 QQEAEDEQQQREQQ 143
+ + E +REQ
Sbjct 2861 LERKKIELAEREQH 2874
> pfa:MAL13P1.176 PfRh2b; reticulocyte binding protein 2, homolog
B
Length=3179
Score = 35.4 bits (80), Expect = 0.16, Method: Composition-based stats.
Identities = 15/57 (26%), Positives = 36/57 (63%), Gaps = 0/57 (0%)
Query 88 EQQKQHQQQRKQEHKQQQQQQLQLRKDEAQADSEAGTQRKGRQQEAEDEQQQREQQQ 144
++Q+Q + Q+++E K+Q+Q++L+ K E E +++ +Q+ + Q+ EQ++
Sbjct 2664 KRQEQERLQKEEELKRQEQERLEREKQEQLQKEEELRKKEQEKQQQRNIQELEEQKK 2720
Score = 30.8 bits (68), Expect = 3.4, Method: Composition-based stats.
Identities = 13/45 (28%), Positives = 29/45 (64%), Gaps = 0/45 (0%)
Query 102 KQQQQQQLQLRKDEAQADSEAGTQRKGRQQEAEDEQQQREQQQMQ 146
K+Q+Q++LQ ++ + + E + K Q + E+E +++EQ++ Q
Sbjct 2664 KRQEQERLQKEEELKRQEQERLEREKQEQLQKEEELRKKEQEKQQ 2708
> hsa:54535 CCHCR1, C6orf18, HCR, MGC126371, MGC126372, SBP; coiled-coil
alpha-helical rod protein 1
Length=835
Score = 31.6 bits (70), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 17/59 (28%), Positives = 35/59 (59%), Gaps = 2/59 (3%)
Query 130 QQEAEDEQQQREQQQMQDMQLLPSVFIFSLQSYGLLRKYS--GVLNLIPATIDANNKGV 186
Q+EA E+ QR +++Q+++ ++ + +LQ GLL +Y +L ++P+ +D V
Sbjct 711 QEEARKEEGQRLARRLQELERDKNLMLATLQQEGLLSRYKQQRLLTVLPSLLDKKKSVV 769
> dre:100331010 upstream stimulatory factor 1-like
Length=2028
Score = 31.6 bits (70), Expect = 2.2, Method: Composition-based stats.
Identities = 22/90 (24%), Positives = 39/90 (43%), Gaps = 2/90 (2%)
Query 83 LETCKEQQKQHQQQRKQEHKQQQQQQLQLRKDEAQADSEAGTQRKGRQQEAEDEQQQREQ 142
+ T ++H + H+Q Q + L++ AQ +S+ Q G + +QQQ ++
Sbjct 1200 MSTVLRPTERHCLTQVSSHEQNQPSVVHLQQGHAQHNSQQSGQNLGGNPYLKHQQQQEQR 1259
Query 143 Q--QMQDMQLLPSVFIFSLQSYGLLRKYSG 170
Q+Q P I S+ LL+ G
Sbjct 1260 HLYQLQHHLTQPESQIHSIHQRNLLQDQHG 1289
Lambda K H
0.317 0.128 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6318090968
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40