bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1227_orf1
Length=152
Score E
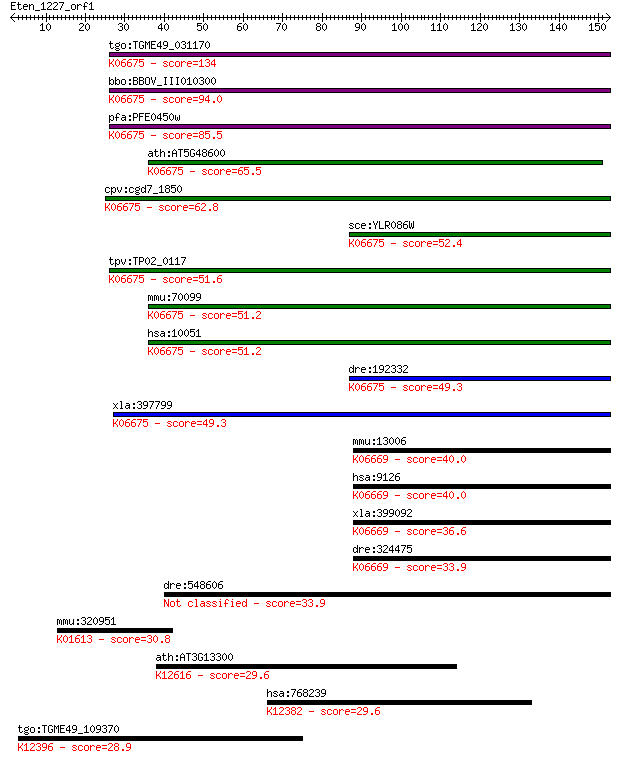
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_031170 chromosome condensation protein, putative ; ... 134 1e-31
bbo:BBOV_III010300 17.m07893; SMC family, C-terminal domain co... 94.0 2e-19
pfa:PFE0450w chromosome condensation protein, putative; K06675... 85.5 7e-17
ath:AT5G48600 ATSMC3; ATSMC3 (ARABIDOPSIS THALIANA STRUCTURAL ... 65.5 7e-11
cpv:cgd7_1850 SMC4'SMC4, chromosomal ATpase with giant coiled ... 62.8 4e-10
sce:YLR086W SMC4; Smc4p; K06675 structural maintenance of chro... 52.4 6e-07
tpv:TP02_0117 hypothetical protein; K06675 structural maintena... 51.6 9e-07
mmu:70099 Smc4, 2500002A22Rik, C79747, SMC-4, Smc4l1; structur... 51.2 1e-06
hsa:10051 SMC4, CAPC, SMC4L1, hCAP-C; structural maintenance o... 51.2 1e-06
dre:192332 smc4, cb788, chunp6901, fb99b10, smc4l1, wu:fb99b10... 49.3 5e-06
xla:397799 smc4, SMC-4, xcap-c; structural maintenance of chro... 49.3 5e-06
mmu:13006 Smc3, Bamacan, Cspg6, HCAP, Mmip1, SMC-3, SmcD; stru... 40.0 0.003
hsa:9126 SMC3, BAM, BMH, CDLS3, CSPG6, HCAP, SMC3L1; structura... 40.0 0.003
xla:399092 smc3, Cspg6, xsmc3; structural maintenance of chrom... 36.6 0.035
dre:324475 smc3, cspg6, im:7142991, wu:fb22e01, wu:fc30d07; st... 33.9 0.20
dre:548606 erc1a, zgc:198416; ELKS/RAB6-interacting/CAST famil... 33.9 0.23
mmu:320951 Pisd, 9030221M09Rik; phosphatidylserine decarboxyla... 30.8 1.6
ath:AT3G13300 VCS; VCS (VARICOSE); nucleotide binding / protei... 29.6 3.5
hsa:768239 PSAPL1, FLJ40379; prosaposin-like 1 (gene/pseudogen... 29.6 3.5
tgo:TGME49_109370 adaptin, putative ; K12396 AP-3 complex subu... 28.9 6.4
> tgo:TGME49_031170 chromosome condensation protein, putative
; K06675 structural maintenance of chromosome 4
Length=1640
Score = 134 bits (336), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 68/129 (52%), Positives = 97/129 (75%), Gaps = 3/129 (2%)
Query 26 FLAASGGFSSYYVVEKPSDAHELFELLRVYELGRCNALALQVLEKELSGRLAEAEAEAAR 85
F+AA GGF S+ VVE+P DA +F++LR LGR N LAL+VLE++L+G + ++AE AR
Sbjct 693 FMAAGGGFCSFLVVEEPQDATVIFQILRQQNLGRVNILALRVLERDLTGFMQRSDAE-AR 751
Query 86 CTDTSAPRLLDLLHFISPRFKIAFFKSVGNTRVAADMEHATHLAYALRQR--VVTLEGGL 143
PRL+DL+ F R+++AF+K+VG+T VA DM+ A+ +AY+ R+R +VTL+GGL
Sbjct 752 AGGFPLPRLIDLISFKKERYRVAFYKAVGDTVVAPDMDEASKIAYSQRRRGGLVTLDGGL 811
Query 144 IELDGRMVG 152
IE+DGRMVG
Sbjct 812 IEVDGRMVG 820
> bbo:BBOV_III010300 17.m07893; SMC family, C-terminal domain
containing protein; K06675 structural maintenance of chromosome
4
Length=1346
Score = 94.0 bits (232), Expect = 2e-19, Method: Composition-based stats.
Identities = 54/128 (42%), Positives = 78/128 (60%), Gaps = 2/128 (1%)
Query 26 FLAASGGFSSYYVVEKPSDAHELFELLRVYELGRCNALALQVLEKELSGRLAEAEAEAA- 84
F+AA+GG VV+ P A ++F+ LR LGRC+ALAL VL +L ++ + +A
Sbjct 602 FMAAAGGQVDVLVVDTPDVASQVFDELRKRNLGRCSALALSVLNNDLRRQMEAFDRNSAD 661
Query 85 RCTDTSAPRLLDLLHFISPRFKIAFFKSVGNTRVAADMEHATHLAYALRQRVVTLEGGLI 144
R TD L+ L+ P +++ FF ++ T +A ++E AT + Y R+RVVTLEG LI
Sbjct 662 RLTD-DVQYLVQLVQPSQPIYRVCFFSALRETLLAPNLEVATKVGYKHRRRVVTLEGELI 720
Query 145 ELDGRMVG 152
E DGRM G
Sbjct 721 EPDGRMSG 728
> pfa:PFE0450w chromosome condensation protein, putative; K06675
structural maintenance of chromosome 4
Length=1708
Score = 85.5 bits (210), Expect = 7e-17, Method: Composition-based stats.
Identities = 51/127 (40%), Positives = 74/127 (58%), Gaps = 1/127 (0%)
Query 26 FLAASGGFSSYYVVEKPSDAHELFELLRVYELGRCNALALQVLEKELSGRLAEAEAEAAR 85
F AS S + VVE P+DA LFE +R +GR N L+L VL K L + + E E
Sbjct 754 FTIASNNCSDFVVVENPNDAVLLFEEVRKANIGRVNVLSLSVLNKNLMTIMLKNE-EIYT 812
Query 86 CTDTSAPRLLDLLHFISPRFKIAFFKSVGNTRVAADMEHATHLAYALRQRVVTLEGGLIE 145
+ RL+DL+ F + ++KI F+ + T +A +E A +AY+ ++RVVT+ G LIE
Sbjct 813 QLLPNVYRLIDLIKFKNDKYKICFYYIIKETLLANSLEQAHVIAYSHKKRVVTINGELIE 872
Query 146 LDGRMVG 152
DGR+ G
Sbjct 873 NDGRICG 879
> ath:AT5G48600 ATSMC3; ATSMC3 (ARABIDOPSIS THALIANA STRUCTURAL
MAINTENANCE OF CHROMOSOME 3); ATP binding / transporter; K06675
structural maintenance of chromosome 4
Length=1241
Score = 65.5 bits (158), Expect = 7e-11, Method: Composition-based stats.
Identities = 46/120 (38%), Positives = 59/120 (49%), Gaps = 13/120 (10%)
Query 36 YYVVEKPSDAHELFELLRVYELGRCNALALQVLEKELSG--RLAEAEAEAARCTDTSAPR 93
Y VVE S A ELLR LG +LEK+ +L E T PR
Sbjct 584 YIVVETTSSAQACVELLRKGNLGFAT---FMILEKQTDHIHKLKEKVK-----TPEDVPR 635
Query 94 LLDLLHFISPRFKIAFFKSVGNTRVAADMEHATHLAYALR---QRVVTLEGGLIELDGRM 150
L DL+ R K+AF+ ++GNT VA D++ AT +AY +RVV L+G L E G M
Sbjct 636 LFDLVRVKDERMKLAFYAALGNTVVAKDLDQATRIAYGGNREFRRVVALDGALFEKSGTM 695
> cpv:cgd7_1850 SMC4'SMC4, chromosomal ATpase with giant coiled
coil regions' ; K06675 structural maintenance of chromosome
4
Length=1366
Score = 62.8 bits (151), Expect = 4e-10, Method: Composition-based stats.
Identities = 45/130 (34%), Positives = 71/130 (54%), Gaps = 7/130 (5%)
Query 25 LFLAASGGFSSYYVVEKPSDAHELFELLRVYELGRCNALALQVLEKELSGRLAEAEAEAA 84
L LA+S VV+ DA E+ +R LGR + + +LEK LS L + +
Sbjct 636 LALASSVPQVENIVVQTTEDAQEVVNYVRKSNLGRISCI---ILEK-LSATLIQNMEKTF 691
Query 85 RCTDTSAPRLLDLLHFISPRFKIAFFKSVGNTRVAADMEHATHLAYALRQ--RVVTLEGG 142
+ + S R DL+ F P+FKIA++ ++ +T + D++ AT ++Y +Q RVVT+ G
Sbjct 692 KAPEGSK-RFFDLVKFKDPKFKIAWYFAMRDTLIVDDLDIATKISYNGKQRWRVVTINGE 750
Query 143 LIELDGRMVG 152
LI+ G M G
Sbjct 751 LIDSSGTMTG 760
> sce:YLR086W SMC4; Smc4p; K06675 structural maintenance of chromosome
4
Length=1418
Score = 52.4 bits (124), Expect = 6e-07, Method: Composition-based stats.
Identities = 26/67 (38%), Positives = 44/67 (65%), Gaps = 1/67 (1%)
Query 87 TDTSAPRLLDLLHFISPRFKIAFFKSVGNTRVAADMEHATHLAYALRQ-RVVTLEGGLIE 145
T + PRL DL+ +P+F AF+ + +T VA +++ A ++AY ++ RVVT++G LI+
Sbjct 757 TPENVPRLFDLVKPKNPKFSNAFYSVLRDTLVAQNLKQANNVAYGKKRFRVVTVDGKLID 816
Query 146 LDGRMVG 152
+ G M G
Sbjct 817 ISGTMSG 823
> tpv:TP02_0117 hypothetical protein; K06675 structural maintenance
of chromosome 4
Length=1398
Score = 51.6 bits (122), Expect = 9e-07, Method: Composition-based stats.
Identities = 37/135 (27%), Positives = 65/135 (48%), Gaps = 19/135 (14%)
Query 26 FLAASGGFSSYYVVEKPSDAHELFELLRVYELGRCNALALQVLEKELSGRLAEAEAEAAR 85
FLA G F Y+VVE P A ++F LR LG+ + +AL V+ + + A++ +
Sbjct 598 FLAICGAFLEYFVVETPEVATKIFTQLRQNNLGKVSCIALSVIS-------SHSNADSGQ 650
Query 86 CTDTSAPRLLDLLHFISPRFKIAFFKSV--------GNTRVAADMEHATHLAYALRQRVV 137
+ L ++ P K ++ + + V +++ A ++ +RVV
Sbjct 651 ----QCGKYGFLNKYVFPNSKDENYEKIVKVINYYTRDVLVVENLDEADEVSRKTNKRVV 706
Query 138 TLEGGLIELDGRMVG 152
T+EG +IE DGR+ G
Sbjct 707 TIEGEVIEKDGRITG 721
> mmu:70099 Smc4, 2500002A22Rik, C79747, SMC-4, Smc4l1; structural
maintenance of chromosomes 4; K06675 structural maintenance
of chromosome 4
Length=1286
Score = 51.2 bits (121), Expect = 1e-06, Method: Composition-based stats.
Identities = 36/119 (30%), Positives = 58/119 (48%), Gaps = 9/119 (7%)
Query 36 YYVVEKPSDAHELFELLRVYELGRCNALALQVLEKELSGRLAEAEAEAARCTDTSAPRLL 95
Y VV+ A E L+ + +G + L + A+ + T + PRL
Sbjct 640 YIVVDSIDTAQECVNFLKKHNIGIATFIGLDKMT-------VWAKKMSKIQTPENTPRLF 692
Query 96 DLLHFISPRFKIAFFKSVGNTRVAADMEHATHLAYA--LRQRVVTLEGGLIELDGRMVG 152
DL+ + + AF+ ++ +T VA +++ AT +AY R RVVTL+G +IE G M G
Sbjct 693 DLVKVKNEEIRQAFYFALRDTLVANNLDQATRVAYQRDRRWRVVTLQGQIIEQSGTMSG 751
> hsa:10051 SMC4, CAPC, SMC4L1, hCAP-C; structural maintenance
of chromosomes 4; K06675 structural maintenance of chromosome
4
Length=1288
Score = 51.2 bits (121), Expect = 1e-06, Method: Composition-based stats.
Identities = 36/119 (30%), Positives = 59/119 (49%), Gaps = 9/119 (7%)
Query 36 YYVVEKPSDAHELFELLRVYELGRCNALALQVLEKELSGRLAEAEAEAARCTDTSAPRLL 95
Y VV+ A E L+ +G + L + + ++ E + T + PRL
Sbjct 642 YIVVDSIDIAQECVNFLKRQNIGVATFIGLDKMAV-WAKKMTEIQ------TPENTPRLF 694
Query 96 DLLHFISPRFKIAFFKSVGNTRVAADMEHATHLAYA--LRQRVVTLEGGLIELDGRMVG 152
DL+ + + AF+ ++ +T VA +++ AT +AY R RVVTL+G +IE G M G
Sbjct 695 DLVKVKDEKIRQAFYFALRDTLVADNLDQATRVAYQKDRRWRVVTLQGQIIEQSGTMTG 753
> dre:192332 smc4, cb788, chunp6901, fb99b10, smc4l1, wu:fb99b10;
structural maintenance of chromosomes 4; K06675 structural
maintenance of chromosome 4
Length=1289
Score = 49.3 bits (116), Expect = 5e-06, Method: Composition-based stats.
Identities = 27/68 (39%), Positives = 40/68 (58%), Gaps = 2/68 (2%)
Query 87 TDTSAPRLLDLLHFISPRFKIAFFKSVGNTRVAADMEHATHLAYA--LRQRVVTLEGGLI 144
T + PRL D++ + AF+ ++ +T VA D+E AT +A+ R RVVTL+G +I
Sbjct 685 TPENIPRLFDMVRVKDESVRPAFYFALRDTLVAKDLEQATRVAFQKDKRWRVVTLQGQII 744
Query 145 ELDGRMVG 152
E G M G
Sbjct 745 EQAGTMTG 752
> xla:397799 smc4, SMC-4, xcap-c; structural maintenance of chromosomes
4; K06675 structural maintenance of chromosome 4
Length=1290
Score = 49.3 bits (116), Expect = 5e-06, Method: Composition-based stats.
Identities = 39/131 (29%), Positives = 66/131 (50%), Gaps = 15/131 (11%)
Query 27 LAASGGFSSYYVVEKPSDAHELFELLRVYELGRCNALAL---QVLEKELSGRLAEAEAEA 83
+++S G + VV+ A E L+ +G + L +V EK L+ ++ E
Sbjct 627 ISSSCGALDHIVVDTIDTAQECVNFLKKQNVGVATFIGLDKMKVWEKGLN-KIQTPE--- 682
Query 84 ARCTDTSAPRLLDLLHFISPRFKIAFFKSVGNTRVAADMEHATHLAYA--LRQRVVTLEG 141
+ PRL D++ + K AF+ ++ +T VA +++ AT +A+ R RVVTL+G
Sbjct 683 ------NIPRLFDMVKVKDEQIKPAFYFALRDTIVANNLDQATRVAFQKDKRWRVVTLQG 736
Query 142 GLIELDGRMVG 152
+IE G M G
Sbjct 737 QIIEQSGTMTG 747
> mmu:13006 Smc3, Bamacan, Cspg6, HCAP, Mmip1, SMC-3, SmcD; structural
maintenace of chromosomes 3; K06669 structural maintenance
of chromosome 3 (chondroitin sulfate proteoglycan 6)
Length=1217
Score = 40.0 bits (92), Expect = 0.003, Method: Composition-based stats.
Identities = 25/70 (35%), Positives = 32/70 (45%), Gaps = 5/70 (7%)
Query 88 DTSAPRLLDLLHFIS-----PRFKIAFFKSVGNTRVAADMEHATHLAYALRQRVVTLEGG 142
DT+ P D + IS PRF AF G T + ME +T LA A +TLEG
Sbjct 597 DTAYPETNDAIPMISKLRYNPRFDKAFKHVFGKTLICRSMEVSTQLARAFTMDCITLEGD 656
Query 143 LIELDGRMVG 152
+ G + G
Sbjct 657 QVSHRGALTG 666
> hsa:9126 SMC3, BAM, BMH, CDLS3, CSPG6, HCAP, SMC3L1; structural
maintenance of chromosomes 3; K06669 structural maintenance
of chromosome 3 (chondroitin sulfate proteoglycan 6)
Length=1217
Score = 40.0 bits (92), Expect = 0.003, Method: Composition-based stats.
Identities = 25/70 (35%), Positives = 32/70 (45%), Gaps = 5/70 (7%)
Query 88 DTSAPRLLDLLHFIS-----PRFKIAFFKSVGNTRVAADMEHATHLAYALRQRVVTLEGG 142
DT+ P D + IS PRF AF G T + ME +T LA A +TLEG
Sbjct 597 DTAYPETNDAIPMISKLRYNPRFDKAFKHVFGKTLICRSMEVSTQLARAFTMDCITLEGD 656
Query 143 LIELDGRMVG 152
+ G + G
Sbjct 657 QVSHRGALTG 666
> xla:399092 smc3, Cspg6, xsmc3; structural maintenance of chromosomes
3; K06669 structural maintenance of chromosome 3 (chondroitin
sulfate proteoglycan 6)
Length=1217
Score = 36.6 bits (83), Expect = 0.035, Method: Composition-based stats.
Identities = 24/70 (34%), Positives = 31/70 (44%), Gaps = 5/70 (7%)
Query 88 DTSAPRLLDLLHFISP-----RFKIAFFKSVGNTRVAADMEHATHLAYALRQRVVTLEGG 142
DT+ P D + IS RF AF G T + ME +T LA A +TLEG
Sbjct 597 DTAYPETNDAIPMISKLRYNLRFDKAFKHVFGKTLICRSMEVSTQLARAFTMDCITLEGD 656
Query 143 LIELDGRMVG 152
+ G + G
Sbjct 657 QVSHRGALTG 666
> dre:324475 smc3, cspg6, im:7142991, wu:fb22e01, wu:fc30d07;
structural maintenance of chromosomes 3; K06669 structural maintenance
of chromosome 3 (chondroitin sulfate proteoglycan
6)
Length=1216
Score = 33.9 bits (76), Expect = 0.20, Method: Composition-based stats.
Identities = 23/70 (32%), Positives = 30/70 (42%), Gaps = 5/70 (7%)
Query 88 DTSAPRLLDLLHFISP-----RFKIAFFKSVGNTRVAADMEHATHLAYALRQRVVTLEGG 142
DT+ P D + IS F AF G T + ME +T LA A +TLEG
Sbjct 597 DTAYPETNDAIPMISKLRYSQNFDKAFKHVFGKTLICRSMEVSTQLARAFTMDCITLEGD 656
Query 143 LIELDGRMVG 152
+ G + G
Sbjct 657 QVSHRGALTG 666
> dre:548606 erc1a, zgc:198416; ELKS/RAB6-interacting/CAST family
member 1a
Length=966
Score = 33.9 bits (76), Expect = 0.23, Method: Composition-based stats.
Identities = 38/114 (33%), Positives = 48/114 (42%), Gaps = 17/114 (14%)
Query 40 EKPSDAHELFELLRVYELGRCNALALQVLEKELSGRLAEAEAEAARCTDTSAPRLLDLLH 99
E+ A ELF L R E E+ R+ EA+ + D S +LL++LH
Sbjct 266 EQERQARELFLLRRTLE--------------EMELRM-EAQRQTLVARDDSVKKLLEMLH 310
Query 100 FISPRFKIAFFKSVGNTRVAADME-HATHLAYALRQRVVTLEGGLIELDGRMVG 152
P K A + TR AD E HA HL L QR L EL R+ G
Sbjct 311 SKGPSAK-ASEEDHERTRRLADAEMHAHHLENLLEQRDKELVALREELHRRLEG 363
> mmu:320951 Pisd, 9030221M09Rik; phosphatidylserine decarboxylase
(EC:4.1.1.65); K01613 phosphatidylserine decarboxylase
[EC:4.1.1.65]
Length=406
Score = 30.8 bits (68), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 13 PLSLLLLLLPLPLFLAASGGFSSYYVVEK 41
P+ L LL P+P+ LA GG++ Y EK
Sbjct 54 PVRSLFLLRPVPILLATGGGYAGYRQYEK 82
> ath:AT3G13300 VCS; VCS (VARICOSE); nucleotide binding / protein
homodimerization; K12616 enhancer of mRNA-decapping protein
4
Length=1344
Score = 29.6 bits (65), Expect = 3.5, Method: Composition-based stats.
Identities = 28/87 (32%), Positives = 37/87 (42%), Gaps = 16/87 (18%)
Query 38 VVEKPSDAHELFELLRVYELGRCNALALQV---LEKEL--------SGRLAEAEAEAARC 86
VV +P D H + V R + + L L +E+ G L A+AE+ RC
Sbjct 393 VVLRPHDGHPVSSATFVTSPERPDHIILITGGPLNREMKIWVSAGEEGWLLPADAESWRC 452
Query 87 TDTSAPRLLDLLHFISPRFKIAFFKSV 113
T T LDL PR + AFF V
Sbjct 453 TQT-----LDLKSSTEPRAEEAFFNQV 474
> hsa:768239 PSAPL1, FLJ40379; prosaposin-like 1 (gene/pseudogene);
K12382 saposin
Length=521
Score = 29.6 bits (65), Expect = 3.5, Method: Composition-based stats.
Identities = 22/67 (32%), Positives = 29/67 (43%), Gaps = 4/67 (5%)
Query 66 QVLEKELSGRLAEAEAEAARCTDTSAPRLLDLLHFISPRFKIAFFKSVGNTRVAADMEHA 125
LE+ S A E DT +P L+ L+ I+P F + GN R A A
Sbjct 319 HALERVCSVMPASITKECIILVDTYSPSLVQLVAKITPEKVCKFIRLCGNRRRA----RA 374
Query 126 THLAYAL 132
H AYA+
Sbjct 375 VHDAYAI 381
> tgo:TGME49_109370 adaptin, putative ; K12396 AP-3 complex subunit
delta-1
Length=1355
Score = 28.9 bits (63), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 22/74 (29%), Positives = 31/74 (41%), Gaps = 2/74 (2%)
Query 3 LSFLLLIFPLPLSLLLLLLPLPLFLAASGGFSSYYVVE--KPSDAHELFELLRVYELGRC 60
L +L ++ LL PLP + +SG + + P AH+L L E GR
Sbjct 1210 LQYLRIVACAANMFLLPQQPLPASVCSSGVLKCLLIAQLYNPDGAHQLQTRLEKVEAGRT 1269
Query 61 NALALQVLEKELSG 74
A L +LSG
Sbjct 1270 ECFAPPSLPADLSG 1283
Lambda K H
0.326 0.142 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3264639800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40