bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1199_orf8
Length=1007
Score E
Sequences producing significant alignments: (Bits) Value
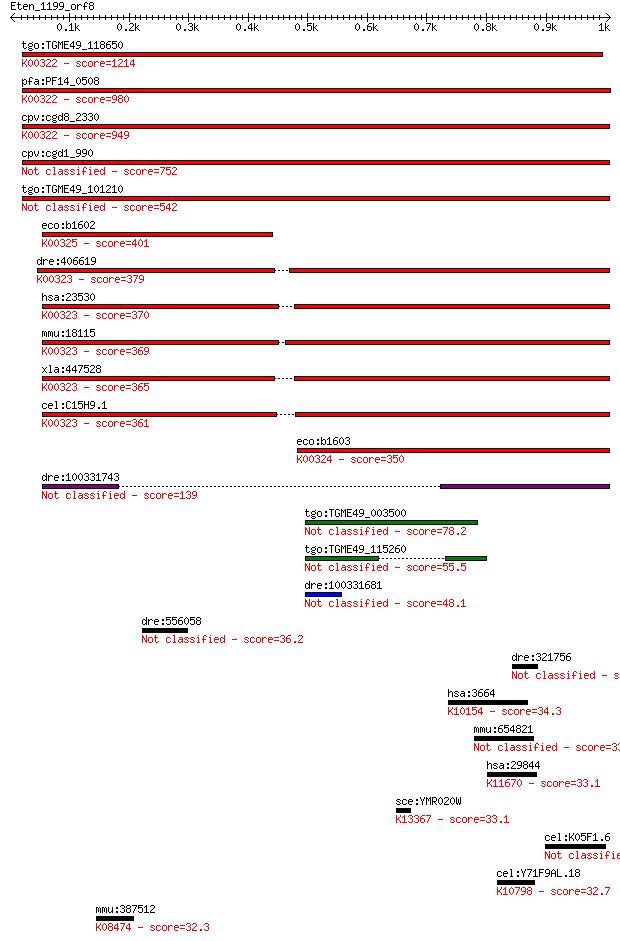
tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00... 1214 0.0
pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative (... 980 0.0
cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alp... 949 0.0
cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alph... 752 0.0
tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, puta... 542 3e-153
eco:b1602 pntB, ECK1597, JW1594; pyridine nucleotide transhydr... 401 7e-111
dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamid... 379 2e-104
hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide t... 370 2e-101
mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide... 369 2e-101
xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydroge... 365 5e-100
cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase fa... 361 8e-99
eco:b1603 pntA, ECK1598, JW1595; pyridine nucleotide transhydr... 350 2e-95
dre:100331743 Nicotinamide Nucleotide Transhydrogenase family ... 139 6e-32
tgo:TGME49_003500 alanine dehydrogenase, putative (EC:1.4.1.1) 78.2 2e-13
tgo:TGME49_115260 alanine dehydrogenase, putative (EC:1.4.1.1) 55.5 1e-06
dre:100331681 MGC83563 protein-like 48.1 2e-04
dre:556058 si:ch211-194c3.5 36.2 0.61
dre:321756 tgif1, Tgif, fa06h05, fb33g11, wu:fa06h05, wu:fb33g... 36.2 0.79
hsa:3664 IRF6, LPS, OFC6, PIT, PPS, VWS, VWS1; interferon regu... 34.3 2.4
mmu:654821 Gcnt7, A330041C17Rik; glucosaminyl (N-acetyl) trans... 33.9 3.2
hsa:29844 TFPT, FB1, INO80F, amida; TCF3 (E2A) fusion partner ... 33.1 5.6
sce:YMR020W FMS1; Fms1p (EC:1.5.3.11); K13367 non-specific pol... 33.1 5.8
cel:K05F1.6 hypothetical protein 33.1 6.3
cel:Y71F9AL.18 pme-1; Poly(ADP-ribose) Metabolism Enzyme famil... 32.7 8.4
mmu:387512 Tas2r135, Tas2r35, mt2r38; taste receptor, type 2, ... 32.3 9.4
> tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00322
NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1013
Score = 1214 bits (3140), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 619/977 (63%), Positives = 775/977 (79%), Gaps = 10/977 (1%)
Query 21 LVGMVAAVVVTFTEAGFGQHYSLFFATAAPAVGLGLYIAQSVNMTEMPQLVALFHSFVGL 80
+ GM AA+++TF+ GFG HY+ FF T AP +G+YIAQSV+M +MPQLVALFHSFVG
Sbjct 34 MTGMGAAILITFSTEGFGGHYAAFFLTVAPPAIVGVYIAQSVSMVQMPQLVALFHSFVGF 93
Query 81 AAVMVGFANFHSPAGVERASSLLRLLEVYAGVFVAGITFTGSVVAAAKLHGSMESRSLRV 140
AAV+VG ++FH+ G E ++ R +E G FVA +TFTGS+VAAAKLH M+ +SL++
Sbjct 94 AAVLVGISSFHAGLG-ESGTNATRAVETAVGEFVAAVTFTGSLVAAAKLHELMDPKSLKI 152
Query 141 PGRHALNTATIAAIGVLGALFCVSSGHFTRMLCLYVNAGLSMWLGFHLVAAIGGADMPVV 200
PGRHA+N T+AA+ V+G FC ++ T+++CLY LS+WLGFHLVA+IGGADMPVV
Sbjct 153 PGRHAINAMTVAAVAVVGITFCATADTTTKIICLYTLITLSLWLGFHLVASIGGADMPVV 212
Query 201 ISLLNSYSGVALAASGFMLDNNLLIIAGALIASSGAILSYIMCKGMNRSLWNVVLGGFEE 260
IS+LNSYSG A AASGFMLDNNLLIIAG+LI SSGAILSYIMC+GMNR LW+VVLGG+E+
Sbjct 213 ISMLNSYSGFATAASGFMLDNNLLIIAGSLIGSSGAILSYIMCRGMNRDLWSVVLGGWED 272
Query 261 AEEVGAASPQGAVQQATADQVADELLAARKVLIVPGYGMAVARCQSELADIAKNLMNCGI 320
A G +G V++ + D VA+E+L A+KVLIVPGYGMAV+RCQS++ADIA+ L + G+
Sbjct 273 APAEGVPGVEGVVREISPDSVAEEVLLAKKVLIVPGYGMAVSRCQSDVADIAQILASRGV 332
Query 321 TVDFGIHPVAGRMPGHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAA 380
V+FGIHPVAGRMPGHMNVLLAEA+V Y+ VKEMSEVN +M YDVV+VVGANDTVNPA+
Sbjct 333 EVEFGIHPVAGRMPGHMNVLLAEANVSYRSVKEMSEVNKQMLEYDVVIVVGANDTVNPAS 392
Query 381 LEPGSKISGMPVIEAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVF 440
LEPG+KI GMPVIE WKA+RV VLKRS+A GYA+I+NPLF L+NTRMLFGNAK+TT+A+F
Sbjct 393 LEPGTKIYGMPVIEVWKAKRVIVLKRSLAPGYAAIDNPLFFLDNTRMLFGNAKDTTTAIF 452
Query 441 ARVNARAEQMPPSAARDDLEAGL--LEFDREERVDPSSWPYPRMAVGVLRDSNG--SVMV 496
A ++ RA + SA DLE G LE R E P++WP P+ VGVL+DSNG + +V
Sbjct 453 ACLSQRAAFVGKSAVFLDLEGGARALEEGRRE-TPPTTWPSPKRTVGVLKDSNGRGTPLV 511
Query 497 PVAPKFVPKLRKLAFRVNVESGAGADAGFTDEEYRRAGAEVLSGPDAVINQSQVLLRVSA 556
+AP+FV +LRK AFRV VESGAGA+A F+D++Y +AGAE++ D VI++S V+L+VS
Sbjct 512 SLAPRFVKRLRKQAFRVIVESGAGAEANFSDDDYVKAGAEIMPNADTVISRSDVILKVSV 571
Query 557 PSPDLVSRIPRDKVLISYLFPSINQQALDMLARQGVTALAVDEVPRVTRAQKLDVKSAMQ 616
PS +L+ RIPR K+LIS++FP N L+++A QG+TALAVDEVPR+TRAQ +DVKS+MQ
Sbjct 572 PSEELIRRIPRGKILISHVFPGQNAPLLELMASQGLTALAVDEVPRITRAQNVDVKSSMQ 631
Query 617 GLQGYRAVIEAFNALPKLSKASISAAGRVEAAKVFVIGAGVAGLQAISTAHGLGAQVFGH 676
GLQGYRAV+EAFNALP+LSK SI+AAGRV+AA+V V+GAGVAGLQAISTAHGL A+VF +
Sbjct 632 GLQGYRAVLEAFNALPRLSKTSITAAGRVDAARVLVLGAGVAGLQAISTAHGLQAEVFAY 691
Query 677 DVRSATREEVESCGGKFIGLRMGEEGEVLGGYAREMGDAYQRAQREMIANTIKHCDVVIC 736
DVR+ATREEVESCGG F+ + + EEGEV GGYAREMG+AY+ AQ++M++ + + DV+IC
Sbjct 692 DVRAATREEVESCGGTFLSVELEEEGEVEGGYAREMGEAYEMAQKQMLSRVVPNVDVIIC 751
Query 737 TAAIHGRPSPKLISRDMLRSMKPGSVVVDLATEFGDVRSGWGGNVEVSPKDDQIVVDGVT 796
TAAIHG+PSPKLIS DML +M+PGSVV+DLATEFGD RS WGGNVE SP D + + GVT
Sbjct 752 TAAIHGKPSPKLISNDMLATMRPGSVVIDLATEFGDRRSNWGGNVEGSPTDGETQIHGVT 811
Query 797 VIGRRRIETRMPIQASELFSMNICNLLEDLGGGSNFRINMDDEVIRGLVAVYQGRNVWQP 856
+IGR RIET+MP QASELFSMN+ NLLE+LGGG NFRI++ +EVI+GL V++GR W P
Sbjct 812 IIGRSRIETQMPTQASELFSMNMSNLLEELGGGENFRIDLTNEVIKGLCCVHEGRVSWAP 871
Query 857 SQPTPVSRTPPRGQMPPPSAPGAPAPEKPGAFAQALASDAFFAMCLVVAAAVVGLLGIVL 916
+P P P + P P P++ Q + SDAFFAM LV AA GLLG+ L
Sbjct 872 PEPLPRRPPTPSPRSSPRLVP----PKEVSLLDQLVTSDAFFAMSLVCTAAFAGLLGVTL 927
Query 917 DPVELKHLTLLGLSLIVGYYCVWAVTPSLHTPLMSVTNALSGVIVIGCMLEYGTAMISGF 976
+EL+ TLL LSLIVGYY VW+VTP+LHTPLMSVTNALSGVI+IG MLEYG + S
Sbjct 928 KALELQQFTLLALSLIVGYYSVWSVTPALHTPLMSVTNALSGVIIIGSMLEYGPSATSAS 987
Query 977 TLLALIGTFLASVNVAG 993
+ AL TF +S+N+AG
Sbjct 988 AICALCATFFSSLNIAG 1004
> pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative
(EC:1.6.1.2); K00322 NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1176
Score = 980 bits (2533), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 487/1050 (46%), Positives = 686/1050 (65%), Gaps = 64/1050 (6%)
Query 21 LVGMVAAVVVTFTEAGFGQHYSLFFATAAPAVGLGLYIAQSVNMTEMPQLVALFHSFVGL 80
+G+VAA+++TF++ GFG Y LF PA+ +G+YIA +V+M +MPQLVALFHSFVGL
Sbjct 128 FIGIVAAIMITFSQVGFGFRYKLFLLIVIPAITIGIYIAHNVSMVQMPQLVALFHSFVGL 187
Query 81 AAVMVGFANFHSPAGVERASSLLRLLEVYAGVFVAGITFTGSVVAAAKLHGSMESRSLRV 140
AA+ VGF+ +HS + S + LLE+Y G F+A I F GS+VAA KL G ++S+SL++
Sbjct 188 AALFVGFSKYHSESFENYEISTIHLLELYVGTFIASIAFIGSLVAAGKLSGILDSKSLKL 247
Query 141 PGRHALNTATIAAIGVLGALFCVSSGHFTRMLCLYVNAGLSMWLGFHLVAAIGGADMPVV 200
+ +N I I +LG F + + +CLY++ + +LGFHL+A+IG ADMPV+
Sbjct 248 QIKKIINILCIVLIIILGYYFVTLKLLYLKSICLYISLIIDSFLGFHLIASIGAADMPVI 307
Query 201 ISLLNSYSGVALAASGFMLDNNLLIIAGALIASSGAILSYIMCKGMNRSLWNVVLGGFEE 260
IS+LNSYSG A A SGF+L NNLLII+GALI SSGAILSYIMC GMNR ++N++LGG+++
Sbjct 308 ISVLNSYSGFATAISGFLLHNNLLIISGALIGSSGAILSYIMCIGMNRDIFNIILGGWDD 367
Query 261 AEEVGAA---------SPQGAVQQATADQVADELLAARKVLIVPGYGMAVARCQSELADI 311
E +G + + + T VA+ L+ A+ ++IVPGYG A+++CQ ELA+I
Sbjct 368 YENMGESIYDQNFIEKKNKQTINSTTNKYVAENLINAKNIIIVPGYGTALSKCQRELAEI 427
Query 312 AKNLMNCGITVDFGIHPVAGRMPGHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVG 371
L + I V F IHPVAGRMPGH+NVLLAEA++PY IVKEM+E+NP +S D+VLVVG
Sbjct 428 CSILTSRNINVTFAIHPVAGRMPGHLNVLLAEANIPYNIVKEMNEINPIISEADIVLVVG 487
Query 372 ANDTVNPAALEPGSKISGMPVIEAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGN 431
AND VNP++L+P SKI GMPVIE WK+++V V KR++ GY++I+NPLF+ NT +LFG+
Sbjct 488 ANDIVNPSSLDPSSKIYGMPVIEVWKSKQVIVFKRTLNTGYSAIDNPLFYFSNTFLLFGD 547
Query 432 AKNTTSAVFARVNARAEQMPPSAARDD--LEAGLLEF-------DREERVDPSS-----W 477
AK+TT+ + +N P + D + +F D D S+ +
Sbjct 548 AKHTTNQILTILNDYVNNKYPDISDQDRHINHDKTQFRYSYSLTDSSHSNDDSTSKEQNY 607
Query 478 PYPRMAVGVLRDSN-----------------------------GSVMVPVAPKFVPKLRK 508
P PR +G+++D N +VP++PKF+PKLR
Sbjct 608 PKPRRVIGLIKDDNVQGGNDLLIEHIQSKVENAPNIKDQKRDVNLSIVPISPKFIPKLRL 667
Query 509 LAFRVNVESGAGADAGFTDEEYRRAGAEVLSGPDAVINQSQVLLRVSAPSPDLVSRIPRD 568
+ FR+ VE G + ++EY + GAEV+S + ++ QS ++L+V P+ + + I +
Sbjct 668 MGFRILVERDIGTNILMQNDEYTKYGAEVVS-RNVILQQSNIILKVDPPTVNFIEEIQNN 726
Query 569 KVLISYLFPSINQQALDML----ARQGVTALAVDEVPRVTRAQKLDVKSAMQGLQGYRAV 624
+LISYL+PSIN LD + + +T LA+DEVPR TRAQKLDV+S+M LQGYRAV
Sbjct 727 TILISYLWPSINYHLLDKMIQDEEKHNITYLAIDEVPRSTRAQKLDVRSSMSNLQGYRAV 786
Query 625 IEAFNALPKLSKASISAAGRVEAAKVFVIGAGVAGLQAISTAHGLGAQVFGHDVRSATRE 684
+EAF LP+ SK+SI+AAG++ AKVFVIGAGVAGLQAI TA LGA V+ HD R AT E
Sbjct 787 LEAFFILPRFSKSSITAAGKINPAKVFVIGAGVAGLQAIITAKSLGAIVYSHDSRLATEE 846
Query 685 EVESCGGKFIGLRMGEEGEVLGGYAREMGDAYQRAQREMIANTIKHCDVVICTAAIHGRP 744
EV+SCGG FI + E G++L + Y + Q + IK CD++IC+A+I G+
Sbjct 847 EVKSCGGIFIRIPTSERGDILNMSTDMNNEEYIKVQSNLFKKIIKKCDILICSASIPGKT 906
Query 745 SPKLISRDMLRSMKPGSVVVDLATEFGDVRSGWGGNVEVSPKDDQIVVDGVTVIGRRRIE 804
SPKL++ +M++ MKPGSV VDL+TEFGD ++ WGGN+E S ++ I+++GV V+GR +IE
Sbjct 907 SPKLVTTEMIKLMKPGSVAVDLSTEFGDKKNNWGGNIECSQSNENILINGVHVLGRDKIE 966
Query 805 TRMPIQASELFSMNICNLLEDLGGGSNFRINMDDEVIRGLVAVYQGRNVWQPSQPT---- 860
MP+QAS+LFSMN+ NLLE++GGG +F I+M++++I+ LV + G ++ P +
Sbjct 967 RNMPMQASDLFSMNMINLLEEMGGGVHFNIDMNNDIIKSLVVIKDGNILYSPDKSVEKLI 1026
Query 861 ---PVSRTPPRGQMPPPSAPGAPAPEKPGAFAQALASDAFFAMCLVVAAAVVGLLGIVLD 917
V + P S P + + SD FF + L+ + L L
Sbjct 1027 KSESVFINEKNQLIEPISEQKIKYPTGLRLTEKFIESDTFFYISLLFVIILTFLAATYLS 1086
Query 918 PVELKHLTLLGLSLIVGYYCVWAVTPSLHTPLMSVTNALSGVIVIGCMLEYGTAMISGFT 977
+L+ L L LS IVGYYCVW+VTP+LHTPLMS+TNALSGVI+IG M+EYG +
Sbjct 1087 QSDLQSLFLFTLSTIVGYYCVWSVTPALHTPLMSMTNALSGVIIIGSMIEYGNGYKDLSS 1146
Query 978 LLALIGTFLASVNVAGGFFVTHRMLKMFQI 1007
+L+++ TFL+SVN++GGF+VT RML MF I
Sbjct 1147 ILSMLATFLSSVNLSGGFYVTKRMLDMFFI 1176
> cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, possible signal peptide
plus 12 transmembrane regions ; K00322 NAD(P) transhydrogenase
[EC:1.6.1.1]
Length=1143
Score = 949 bits (2453), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 491/1011 (48%), Positives = 679/1011 (67%), Gaps = 38/1011 (3%)
Query 21 LVGMVAAVVVTFTEAGFGQHYSLFFATAAPAVGLGLYIAQSVNMTEMPQLVALFHSFVGL 80
+VG++ A+ TF F ++ +F A A+ +G+ I+ V+M +PQLVALFHSFVGL
Sbjct 123 IVGIICAIAATFLAPTFTMNWIMFIVPFALAIIIGIPISHCVSMVNIPQLVALFHSFVGL 182
Query 81 AAVMVGFANFHSP-AGVERASSLLRLLEVYAGVFVAGITFTGSVVAAAKLHGSMESRSLR 139
AA+++ FAN +P E S + +E++ G ++ ITFTGS+VAA KLH S +L+
Sbjct 183 AAMLISFANLWTPFQSSEEEFSAVHAIEMFIGEAISAITFTGSIVAAGKLHEIFPSGALK 242
Query 140 VPGRHALNTATIAAIGVLGALFCVSSGHFTRMLCLYVNAGLSMWLGFHLVAAIGGADMPV 199
+PGRH LN +A + LG++F + + + R +Y N+ LSM LG HLVA+IGGADMPV
Sbjct 243 LPGRHFLNALMVAGLIALGSVFIIITDYANRTYLMYGNSLLSMLLGIHLVASIGGADMPV 302
Query 200 VISLLNSYSGVALAASGFMLDNNLLIIAGALIASSGAILSYIMCKGMNRSLWNVVLGGFE 259
IS+LNSYSG A + +GF++++ +LII GALI SSGAILS+IMC GMNRSL NV+LGG+E
Sbjct 303 AISMLNSYSGWATSFTGFLINSKMLIICGALIGSSGAILSHIMCLGMNRSLLNVMLGGWE 362
Query 260 ---EAEEVGAASPQGAVQQATADQVADELLAARKVLIVPGYGMAVARCQSELADIAKNLM 316
++ E A Q V + A +VA +LL+A+KVLIVPGYGMAV+R Q ++A I L
Sbjct 363 SNGDSNEAQALGDQSDVNKTNAMKVARDLLSAKKVLIVPGYGMAVSRSQQDVASIVNALR 422
Query 317 NCGITVDFGIHPVAGRMPGHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTV 376
I V+F IHPVAGRMPGHMNVLLAEADVPY IVKEM +VNP M S+DVVLV+GANDTV
Sbjct 423 LRDIYVEFAIHPVAGRMPGHMNVLLAEADVPYSIVKEMEDVNPHMESFDVVLVIGANDTV 482
Query 377 NPAALEPGSKISGMPVIEAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTT 436
NP ALE SKI+GMPVIE W+A +V V KRS+ GYA+I+NPLF ++N M+FGNAK++
Sbjct 483 NPLALEKDSKINGMPVIEVWRASKVIVSKRSLGKGYAAIDNPLFFMKNVEMIFGNAKDSM 542
Query 437 SAVFARVNARAEQMPPSAARDDLEAGLLEFDREERVDPSSWPYPRMAVGVLR-DSNGSVM 495
+ + + + + + DD E ++ + D +P P M +GVL+ D +
Sbjct 543 INILQNIISISPEQKKANLNDDQET--IDGTTASQEDNEEYPTPTMTIGVLKEDLPSEKL 600
Query 496 VPVAPKFVPKLRKLAFRVNVESGAGADAGFTDEEYRRAGAEVLSGPDAVINQSQVLLRVS 555
V +AP FV KLRKL FRV VESGAG + F D++Y A +++ V+++S V+++V
Sbjct 601 VAIAPNFVKKLRKLGFRVLVESGAGTKSQFDDQKYENASCTIMATRQDVVSRSDVIVKVQ 660
Query 556 APSPDLVSRIPRDKVLISYLFPSINQQALDMLARQGVTALAVDEVPRVTRAQKLDVKSAM 615
P+ + +S++ + L+SY++P+ N L+ LA++GVT +A+DEVPR TRAQKLD++S+M
Sbjct 661 KPTDEEISQMKSGQTLVSYIWPAQNPSLLESLAKKGVTTIALDEVPRTTRAQKLDIRSSM 720
Query 616 QGLQGYRAVIEAFNALPKLSKASISAAGRVEAAKVFVIGAGVAGLQAISTAHGLGAQVFG 675
L GYRAVIEAF LPKLSK+SI+AAGRV+AA+VFVIGAGVAGLQAI+TA LGA V+
Sbjct 721 SNLAGYRAVIEAFVQLPKLSKSSITAAGRVDAARVFVIGAGVAGLQAIATAKNLGADVYA 780
Query 676 HDVRSATREEVESCGGKFIGLRMGEEGEVLGGYAREMGDAYQRAQREMIANTIKHCDVVI 735
D R+ATREEVES G KF+ + + E+G+ GYA+ M Y +AQ ++ + I+ CDVVI
Sbjct 781 SDTRTATREEVESLGAKFVTVDIKEDGDSGSGYAKVMSPEYLKAQSKLYSKMIRSCDVVI 840
Query 736 CTAAIHGRPSPKLISRDMLRSMKPGSVVVDLATEFGDVRSGWGGNVEVSPKDDQIVVD-- 793
TA I G+PSPK+I+R+M+ SMKPGSV+VD+A E D SGWGGN E++ K DQI +D
Sbjct 841 TTALIPGKPSPKIITREMVNSMKPGSVIVDMAAEMADTASGWGGNCEIT-KKDQIYLDEK 899
Query 794 -GVTVIGRRRIETRMPIQASELFSMNICNLLEDLGGGSNFRINMDDEVIRGLVAVYQGRN 852
GVT+IG + + MP QASELFSMN+ ++LE+LGG +F ++M D++++ +V +
Sbjct 900 SGVTIIGLTNLPSTMPSQASELFSMNVVHMLEELGGAEHFSVDMKDDLLKEMVVTINEKV 959
Query 853 VWQPSQPTPVSRTPPRGQMPPPSAPGAPAPEK--------------PGAFAQALASDAFF 898
+ PV + PP PP S + + + S+ F
Sbjct 960 TY-----VPVDKRPP----PPKSESSNSSSSSSSSSSSTETSHRSFSSIMERVIYSNVSF 1010
Query 899 AMCLVVAAAVVGLLGIVLDPVELKHLTLLGLSLIVGYYCVWAVTPSLHTPLMSVTNALSG 958
+ ++ V LG ++D L ++ + LS+IVGYYC+W VTPSLHTPLMSVTNALSG
Sbjct 1011 GFFVFISVLVSIGLGYIIDHDTLGNILVFSLSVIVGYYCIWNVTPSLHTPLMSVTNALSG 1070
Query 959 VIVIGCMLEYGTAMI----SGFTLLALIGTFLASVNVAGGFFVTHRMLKMF 1005
+I+IG MLE G ++ ++ L + L+S+N+ GGF+VT RML MF
Sbjct 1071 IIIIGAMLECGPVILFTDFQVYSFLLFLAMLLSSINIIGGFYVTTRMLYMF 1121
> cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, 12 transmembrane domain
(EC:1.6.1.2)
Length=1147
Score = 752 bits (1942), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 416/1071 (38%), Positives = 618/1071 (57%), Gaps = 102/1071 (9%)
Query 21 LVGMVAAVVVTFTEAGFGQHYSLFFATAAPAVGLGLYIAQSVNMTEMPQLVALFHSFVGL 80
VGM+ A++V+ +E GFG HY +FF + +G+YIA + +MPQLVA+FHS VGL
Sbjct 91 FVGMMVAILVSLSEEGFGNHYFVFFLATIVSGAVGVYIADKTQLIKMPQLVAIFHSLVGL 150
Query 81 AAVMVGFANFHSPAGVERASSLLRLLEVYAGVFVAGITFTGSVVAAAKLHGSMESRSLRV 140
AA+ V ++ F+S G++ +LR +E++ G + ITF GSV+AA KL + S+S+++
Sbjct 151 AALFVSYSYFYSSIGMKEGIYVLRRMELFVGGVMGMITFVGSVIAALKLDDIIPSKSVKI 210
Query 141 PGRHALNTATIAAIGVLGALFCVSSGHFTRMLCLYVNAGLSMWLGFHLVAAIGGADMPVV 200
P ++ + + +I + G FCVS+ L+ LS G ++ +IGGADMPV+
Sbjct 211 PLKNVSLSILLFSICMTGLSFCVSNNELVITTNLHCGMILSALFGIFMIISIGGADMPVI 270
Query 201 ISLLNSYSGVALAASGFMLDNNLLIIAGALIASSGAILSYIMCKGMNRSLWNVVLGGFEE 260
IS+LNSYSG + A +GF+LDN+LLII+GALI SSGAILSYIMCKGMNR +V+LGGF++
Sbjct 271 ISMLNSYSGWSTAITGFLLDNSLLIISGALIGSSGAILSYIMCKGMNRDFVSVLLGGFDQ 330
Query 261 AEEVGAASPQGAVQQATADQVADELLAARKVLIVPGYGMAVARCQSELADIAKNLMNCGI 320
EE ++ + A+ L + K+LIVPGYGMAV++CQ ++DI K L +
Sbjct 331 -EETEKMVGDTKCYTTSSKETAEILAESSKILIVPGYGMAVSKCQDIISDIIKELESRST 389
Query 321 TVDFGIHPVAGRMPGHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAA 380
V+ GIHPVAGRMPGHMNVLLAE+ VP +IVKEM VN M YD+VLVVGAND VNPAA
Sbjct 390 IVEIGIHPVAGRMPGHMNVLLAESGVPSRIVKEMDAVNNCMHEYDLVLVVGANDIVNPAA 449
Query 381 LEPGSKISGMPVIEAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVF 440
L+P SKISGMPVI WKA++V V KRS+A GYA IEN LF E TRML G+++ T V+
Sbjct 450 LDPQSKISGMPVINVWKAKQVVVSKRSLAYGYACIENELFTCERTRMLLGDSRETLQQVY 509
Query 441 A------------RVNAR-AEQMPPSAARDDLEAGLLEFDREERVDPSSWPYPRMAVG-- 485
+++R E++ D++ LL + + S R
Sbjct 510 KILKGCAGFVPRQYIDSRVTEEIEEVEDTDEVTTALLSSKQRKSSKTSGNVSSRSKKAGG 569
Query 486 -----------------------VLRDSNGSVMVPVAPKFVPKLRKLAFRV----NVESG 518
++ + + +++ P+ P V K R+ + V +V +
Sbjct 570 AETETEKVSMSANKGSKKIALLPIVSEQDKTLLFPIPPSKVYKFREKGYEVAISRDVLTS 629
Query 519 AGADAGFTDEEYRRAGAEVLSGPDAVINQSQVLLRVSAPSPDLVSRIPRDKVLISYLFPS 578
+ F++E Y R+GA V + +I + V+++++ PS + + + LI + S
Sbjct 630 SFQSKYFSEEIYMRSGALVYKNIEELIKECDVVIKMARPSEKEAKCMKQGQFLICNMHIS 689
Query 579 INQ-------QALDMLARQGVTALAVDEVPRVTRAQKLDVKSAMQGLQGYRAVIEAFNAL 631
Q Q L L + GVT +A+DEVPR +RAQ +D+++ + GYRAV++AFN L
Sbjct 690 QYQDKENSQDQLLASLTKNGVTVIALDEVPRTSRAQTMDIRTTTSTISGYRAVVDAFNYL 749
Query 632 PKLSKASISAAGRVEAAKVFVIGAGVAGLQAISTAHGLGAQVFGHDVRSATREEVESCGG 691
P++SK+ SAAG VE + V VIGAGVAGLQAI+TA +G +VF D RS ++EE ESCG
Sbjct 750 PRISKSISSAAGNVEESTVLVIGAGVAGLQAIATAKSMGTKVFAMDSRSTSKEEAESCGA 809
Query 692 KFIGLRMGEEGEVLGGYAREMGDAYQRAQREMIANTIKHCDVVICTAAIHGRPSPKLISR 751
+FI ++ EGE L + + QR++I + D+VI +A G P LI++
Sbjct 810 RFI--QVPSEGESL------RKEEILKKQRDLIEKYLCISDIVITSACKPGEECPILITK 861
Query 752 DMLRSMKPGSVVVDLATEFGDVRSGWGGNVEVSPKDDQI--VVDGVTVIGRRRIETRMPI 809
+ +R MK GSV+VDL +EF GGN E++ KD V G T+IG+ MP+
Sbjct 862 EAVRKMKSGSVIVDLCSEF-------GGNCELTQKDRTFSDVQSGTTIIGKCNYVFSMPL 914
Query 810 QASELFSMNICNLLEDLGGGSN-FRINMDDEVIRGLVAVYQGRNVWQPSQPTPVSRTPPR 868
Q+SELFS N+ +L+ +LG S+ F+ ++++E+I + ++ + +W+P + +
Sbjct 915 QSSELFSGNLLSLISELGPTSDRFKCDLNNEIISKMCVAHENKMLWRPFTEQNQEKHENQ 974
Query 869 GQMPPPS--------------APGAPAPEKPGAFAQALASDAF----------------- 897
+ S K F D +
Sbjct 975 EMLEKKSLLENISKQSTLISIKDEDTTTSKTSVFCSKTEIDGYLQKVMKEFKNFDKQLNG 1034
Query 898 ---FAMCLVVAAAVVGLLGIVLDPVELKHLTLLGLSLIVGYYCVWAVTPSLHTPLMSVTN 954
F +++A +LG+ + ++++++ +S ++GYYCVW V P LHTPLMSVTN
Sbjct 1035 GVNFYAGVILATLFFTVLGLTMTTIQIQNIFSFIISTMLGYYCVWDVDPKLHTPLMSVTN 1094
Query 955 ALSGVIVIGCMLEYGTAMISGFTLLALIGTFLASVNVAGGFFVTHRMLKMF 1005
ALSGVI+IG M++YG ++ TL+++ TFLAS+N GGF+VT++ML +F
Sbjct 1095 ALSGVIIIGSMMQYGNQTVTYTTLMSMFSTFLASINTFGGFYVTNKMLTLF 1145
> tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, putative
(EC:1.6.1.2)
Length=1165
Score = 542 bits (1396), Expect = 3e-153, Method: Compositional matrix adjust.
Identities = 411/1178 (34%), Positives = 583/1178 (49%), Gaps = 216/1178 (18%)
Query 21 LVGMVAAVVVTFTEAG-FGQHYSLFFATAAPAVGLGLYIAQSVNMTEMPQLVALFHSFVG 79
++GM+ AV+ T + + +FF P L + +A V MT +PQLV ++F G
Sbjct 7 VIGMLVAVIGTLVSPFVYDRALWIFFVVCVPPAVLAVIVAMLVKMTSIPQLVGALNAFGG 66
Query 80 LAAVMVGFANFHSPAGVERASSLLRL-------LEVYAGVF------VAGITFTGSVVAA 126
LAA + FA + SP V + ++ + +VY VF + ITFTGS+VA
Sbjct 67 LAATLESFALYFSPYEVAKQEAMQSVGETRYLQFQVYQTVFYLIGASIGMITFTGSLVAC 126
Query 127 AKLHGSMESRSLRVPGR------HALNTATIAAIGVLGALFCVSSGHFTRMLCLYVNAGL 180
KL G + S+ +P R A+ + L V G LC + L
Sbjct 127 GKLSGWIASKPRVMPLRCFWMPVVLALLLAAGAVAGVLGLDNVPVGTVCLSLCTF----L 182
Query 181 SMWLGFHLVAAIGGADMPVVISLLNSYSGVALAASGFMLDNNLLIIAGALIASSGAILSY 240
S G V AIGGADMPVVIS+LNS SG A +G L N+++IIAGA + +SG ILSY
Sbjct 183 SAVYGVCFVLAIGGADMPVVISVLNSGSGWAGVFAGLTLQNSIIIIAGAFVGASGIILSY 242
Query 241 IMCKGMNRSLWNVVLGGF-EEAEEVGAASPQGAVQQATADQVADELLAARKVLIVPGYGM 299
+MC MNRSL NV+LGGF + +E A + +G + AT++QVA LLA++ V+IVPGYGM
Sbjct 243 VMCTAMNRSLCNVLLGGFGDTGKESNAQAFEGEAKIATSEQVAAYLLASQSVIIVPGYGM 302
Query 300 AVARCQSELADIAKNLMNCGITVDFGIHPVAGRMPGHMNVLLAEADVPYKIVKEMSEVNP 359
AV+ Q + ++ + L G V F IHPVAGR+PGHMNVLLAEA+VPY IV M E+N
Sbjct 303 AVSHAQFAVHELTQQLRERGTQVRFAIHPVAGRLPGHMNVLLAEANVPYDIVLSMDEINE 362
Query 360 EMSSYDVVLVVGANDTVNPAALE-PGSKISGMPVIEAWKARRVFVLKRSMAAGYASIENP 418
+ D V+++GANDTVNPAA PG I GMPV+E WKA++V VLKRSM GYA ++NP
Sbjct 363 DFPQTDTVIIIGANDTVNPAAQNAPGCPIYGMPVLEVWKAKKVIVLKRSMRVGYAGVDNP 422
Query 419 LFHLENTRMLFGNAKNTTSAVFARVNARAEQMPPSAARDDLE-------------AGLLE 465
LF N+ ML G+AK + + + + + P A+R E AG
Sbjct 423 LFFYPNSEMLLGDAKASLQTLLTDMQDKLDAHPIEASRLSSETQTNKSARIAVNVAGDKS 482
Query 466 FDREE-----------------------------RVDP--SSWPYPRMAVGVLRDSNG-S 493
+E+ V P S P P VGVL++
Sbjct 483 AKQEKPHGAAADKPDATETAGTVGVTVTGSGAPGEVPPVAESAP-PLFFVGVLKERRPLE 541
Query 494 VMVPVAPKFVPKLRKLAFRVNVESGAGADAGFTDEEYRRAGAEVLSGPDAVINQSQVLLR 553
V + P V KLR+L +E AG AGF DE+Y +GA + + V+ S+V+++
Sbjct 542 RRVAMTPSVVKKLRELQLGCIIEKQAGVSAGFLDEQYMESGALIADTAEEVVRLSRVVVK 601
Query 554 VSAPSPDLV------------SRIPRDKVLISYLFPSINQQALDMLARQGVTA----LAV 597
V+ D V SR + L + P+ N++ ++ + +G L+
Sbjct 602 VTTFECDEVALAKPQNDDEQSSRESTEPPLPALATPA-NKEDTELYSVEGNGKGHRLLSS 660
Query 598 DEV-------PRVTRAQKLDVKSAMQGLQGYRAVIEA---------------FNALPKLS 635
++V P T ++ SA +G +G + ++A + LP+L+
Sbjct 661 EKVFVAGFVGPN-TATSEVSADSAPEGEKGKLSPVDALLRSALAIPQVTLISLDVLPRLT 719
Query 636 KA--------------------SISAAGRV-----------EAAKVFVIGAGVAGLQAIS 664
+ +IS GR+ AKV VIGAGVAGLQAI
Sbjct 720 ISQKMDVLSSTAKLAGYRAVVEAISHFGRLMGPEITSAGKYPPAKVLVIGAGVAGLQAIG 779
Query 665 TAHGLGAQVFGHDVRSATREEVESCGGKFIGLRM---GEEGEVL-GGYAREMGDAYQRAQ 720
AH LGA V G DVR +E+VES GG+F+ + GE G GGYA+ M + + R +
Sbjct 780 DAHRLGADVRGFDVRLECKEQVESMGGEFLAMTFEGPGESGRATAGGYAKPMSEEFLRKE 839
Query 721 REMIANTIKHCDVVICTAAIHGRPSPKLISRDMLRSMKPGSVVVDLATEFGDVRSGWGGN 780
E+ A + D++I TA++ GRP+PKLI ++ + +MK GSV+VDLA + GGN
Sbjct 840 MELFAEQCREIDILITTASLPGRPAPKLIKKEHVDTMKAGSVIVDLA-------APTGGN 892
Query 781 VEVSPKDDQIVVDG-VTVIGRRRIETRMPIQASELFSMNICNLLEDLGGGSNFRINMDDE 839
VEV+ + + +G VTV+G + +RM QASE+F+ NI NLLE +G G +F +N DDE
Sbjct 893 VEVTRPGETYLYNGKVTVVGWTDLPSRMAPQASEMFARNIFNLLEHMGAGRDFSVNFDDE 952
Query 840 VIRGLVAVYQGRNVWQP--SQPTPVSRTPPRGQMPPPSAPGAPAPEKPGAFAQALASDAF 897
+IR + GR + P + P + + + AP E A S+
Sbjct 953 IIRAITVARHGRKTYPPPRKESDPENTNAGSALLTGSNTDAAPVEE------SAQRSENL 1006
Query 898 FAMC----------LVVAAAVVGLLGIVLD---------PVELKHLTLLGLSLIVGYYCV 938
+ + +V ++ +L I P L + L+ VGY V
Sbjct 1007 WHLVQKRMRNRWRGIVAPLDILTILVIAAFTALFAATAPPSFPPLLFIFFLACFVGYLLV 1066
Query 939 WAVTPSLHTPLMSVTNALSGVIVIGCML------------------------------EY 968
W V P+LHTPLMSVTNA+SG +++G ML
Sbjct 1067 WNVAPALHTPLMSVTNAISGTVLVGGMLGISARAYQTLDKESICPHNHLGPLEPFYCHSR 1126
Query 969 GTAMISGFTLLALIGTFLASVNVAGGFFVTHRMLKMFQ 1006
GTA I+ L I +AS+NV GGF VT RML MF+
Sbjct 1127 GTAAIT----LNAIAIAVASMNVFGGFAVTQRMLNMFR 1160
> eco:b1602 pntB, ECK1597, JW1594; pyridine nucleotide transhydrogenase,
beta subunit (EC:1.6.1.2); K00325 NAD(P) transhydrogenase
subunit beta [EC:1.6.1.2]
Length=462
Score = 401 bits (1031), Expect = 7e-111, Method: Compositional matrix adjust.
Identities = 208/390 (53%), Positives = 273/390 (70%), Gaps = 3/390 (0%)
Query 54 LGLYIAQSVNMTEMPQLVALFHSFVGLAAVMVGFANF-HSPAGVERASSLLRLLEVYAGV 112
+G+ +A+ V MTEMP+LVA+ HSFVGLAAV+VGF ++ H AG+ + L EV+ G+
Sbjct 70 IGIRLAKKVEMTEMPELVAILHSFVGLAAVLVGFNSYLHHDAGMAPILVNIHLTEVFLGI 129
Query 113 FVAGITFTGSVVAAAKLHGSMESRSLRVPGRHALNTATIAAIGVLGALFCVSSGHFTRML 172
F+ +TFTGSVVA KL G + S+ L +P RH +N A + +L +F + ++L
Sbjct 130 FIGAVTFTGSVVAFGKLCGKISSKPLMLPNRHKMNLAALVVSFLLLIVFVRTDSVGLQVL 189
Query 173 CLYVNAGLSMWLGFHLVAAIGGADMPVVISLLNSYSGVALAASGFMLDNNLLIIAGALIA 232
L + +++ G+HLVA+IGGADMPVV+S+LNSYSG A AA+GFML N+LLI+ GAL+
Sbjct 190 ALLIMTAIALVFGWHLVASIGGADMPVVVSMLNSYSGWAAAAAGFMLSNDLLIVTGALVG 249
Query 233 SSGAILSYIMCKGMNRSLWNVVLGGF-EEAEEVGAASPQGAVQQATADQVADELLAARKV 291
SSGAILSYIMCK MNRS +V+ GGF + G G ++ TA++ A+ L + V
Sbjct 250 SSGAILSYIMCKAMNRSFISVIAGGFGTDGSSTGDDQEVGEHREITAEETAELLKNSHSV 309
Query 292 LIVPGYGMAVARCQSELADIAKNLMNCGITVDFGIHPVAGRMPGHMNVLLAEADVPYKIV 351
+I PGYGMAVA+ Q +A+I + L GI V FGIHPVAGR+PGHMNVLLAEA VPY IV
Sbjct 310 IITPGYGMAVAQAQYPVAEITEKLRARGINVRFGIHPVAGRLPGHMNVLLAEAKVPYDIV 369
Query 352 KEMSEVNPEMSSYDVVLVVGANDTVNPAAL-EPGSKISGMPVIEAWKARRVFVLKRSMAA 410
EM E+N + + D VLV+GANDTVNPAA +P S I+GMPV+E WKA+ V V KRSM
Sbjct 370 LEMDEINDDFADTDTVLVIGANDTVNPAAQDDPKSPIAGMPVLEVWKAQNVIVFKRSMNT 429
Query 411 GYASIENPLFHLENTRMLFGNAKNTTSAVF 440
GYA ++NPLF ENT MLFG+AK + A+
Sbjct 430 GYAGVQNPLFFKENTHMLFGDAKASVDAIL 459
> dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P)
transhydrogenase [EC:1.6.1.2]
Length=1079
Score = 379 bits (974), Expect = 2e-104, Method: Compositional matrix adjust.
Identities = 207/407 (50%), Positives = 276/407 (67%), Gaps = 13/407 (3%)
Query 47 TAAPAVG--LGLYIAQSVNMTEMPQLVALFHSFVGLAAVMVGFANF---HSPAGVERASS 101
+AA AVG GL IA+ + ++++PQLVA FHS VGLAAV+ A + + + A++
Sbjct 673 SAAMAVGGTAGLTIAKKIQISDLPQLVAAFHSLVGLAAVLTCVAEYMVEYPHFATDPAAN 732
Query 102 LLRLLEVYAGVFVAGITFTGSVVAAAKLHGSMESRSLRVPGRHALNTATIAAIGVLGAL- 160
L +++ Y G ++ G+TF+GS+VA KL G + S L +PGRHALN AT+ A V G +
Sbjct 733 LTKIV-AYLGTYIGGVTFSGSLVAYGKLQGLLNSAPLMLPGRHALN-ATLMAASVGGMIP 790
Query 161 FCVSSGHFTRMLCLYVNAGLSMWLGFHLVAAIGGADMPVVISLLNSYSGVALAASGFMLD 220
+ + + T + CL + LS +G L AAIGGADMPVVI++LNSYSG AL A GF+L+
Sbjct 791 YMLDPSYTTGITCLGSVSALSAVMGLTLTAAIGGADMPVVITVLNSYSGWALCAEGFLLN 850
Query 221 NNLLIIAGALIASSGAILSYIMCKGMNRSLWNVVLGGFEEAEEVGAASP---QGAVQQAT 277
NNLL I GALI SSGAILSYIMC MNRSL NV+LGG+ + G P G +
Sbjct 851 NNLLTIVGALIGSSGAILSYIMCVAMNRSLANVILGGYGTSS-TGTGKPMEITGTHTEVN 909
Query 278 ADQVADELLAARKVLIVPGYGMAVARCQSELADIAKNLMNCGITVDFGIHPVAGRMPGHM 337
DQ D + A ++IVPGYG+ A+ Q +AD+ K+L + G V FGIHPVAGRMPG +
Sbjct 910 VDQTVDLIKEAHNIIIVPGYGLCAAKAQYPIADLVKSLTDQGKKVRFGIHPVAGRMPGQL 969
Query 338 NVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAALE-PGSKISGMPVIEAW 396
NVLLAEA VPY +V EM E+N + D+VLV+GANDTVN AA E P S I+GMPV+E W
Sbjct 970 NVLLAEAGVPYDVVLEMDEINEDFPETDLVLVIGANDTVNSAAQEDPNSIIAGMPVLEVW 1029
Query 397 KARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARV 443
K+++V V+KRS+ GYA+++NP+F+ NT ML G+AK T A+ A+V
Sbjct 1030 KSKQVVVMKRSLGVGYAAVDNPIFYKPNTSMLLGDAKKTCDALSAKV 1076
Score = 296 bits (759), Expect = 2e-79, Method: Compositional matrix adjust.
Identities = 208/562 (37%), Positives = 299/562 (53%), Gaps = 45/562 (8%)
Query 470 ERVDPSSWPYPRMAVGVLRDS-NGSVMVPVAPKFVPKLRKLAFRVNVESGAGADAGFTDE 528
R+ PY ++ VGV ++ V ++P V L K F V VESGAG A F+D+
Sbjct 41 NRLTSPGIPYKQLTVGVPKEIFQNERRVAISPAGVEALIKQGFNVVVESGAGESAKFSDD 100
Query 529 EYRRAGAEVLSGPDAVINQSQVLLRVSAP--SPDL----VSRIPRDKVLISYLFPSINQQ 582
Y +AGA + D + S VLL+V AP +P L S + L+S+++P+ N +
Sbjct 101 MYTKAGATIRDVKD--VFSSDVLLKVRAPMLNPTLGVHEASLMSEGATLVSFIYPAQNPE 158
Query 583 ALDMLARQGVTALAVDEVPRVTRAQKLDVKSAMQGLQGYRAVIEAFNALPKLSKASISAA 642
+D L+++ T LA+D+VPRVT AQ D S+M + GY+AV+ A N + I+AA
Sbjct 159 LMDTLSQRKATVLAMDQVPRVTIAQGYDALSSMANIAGYKAVVLAANNFGRFFTGQITAA 218
Query 643 GRVEAAKVFVIGAGVAGLQAISTAHGLGAQVFGHDVRSATREEVESCGGKFIGLRMGEEG 702
G+V AKV +IG GVAGL A +A +GA V G D R+A E+ +S G + + + + E G
Sbjct 219 GKVPPAKVLIIGGGVAGLAAAGSARAMGAIVRGFDTRAAALEQFKSLGAEPLEVDIKESG 278
Query 703 EVLGGYAREMGDAYQRAQREMIANTIKHCDVVICTAAIHGRPSPKLISRDMLRSMKPGSV 762
E GGYA+EM + A+ ++ A D++I TA I GR +P LI+++M+ +MK GSV
Sbjct 279 EGQGGYAKEMSKEFIEAEMKLFAKQCLDVDIIITTALIPGRKAPVLITKEMVETMKDGSV 338
Query 763 VVDLATEFGDVRSGWGGNVEVSPKDDQIVVDGVTVIGRRRIETRMPIQASELFSMNICNL 822
VVDLA E GGN+E + + V GV +G I +R+P QAS L+S NI L
Sbjct 339 VVDLAAE-------AGGNIETTVPGELSVHKGVIHVGYTDIPSRLPTQASTLYSNNITKL 391
Query 823 LEDLGGG---------SNFRINMDDEVIRGLVAVYQGRNVWQPSQPTPVSRTPPRGQMPP 873
+ + + F D VIRG V + G+ ++ QP V P P
Sbjct 392 IRAISPDKETFYFDVKNEFDFGTMDHVIRGSVVMQDGKVLFPAPQPQNVPVAAP----PK 447
Query 874 PSAPGAPAPEKPGAFAQALASDAFFAMCLVVAAAVVGLLGIVL-------DPVELKHLTL 926
EK A + F L A G LG + + + +T
Sbjct 448 QKTVQELQKEKASAVSP-------FRATLTTAGVYTGGLGTAIGLGLCAPNAAFTQMVTT 500
Query 927 LGLSLIVGYYCVWAVTPSLHTPLMSVTNALSGVIVIGCMLEYGTAMISGFT--LLALIGT 984
GL+ IVGY+ VW VTP+LH+PLMSVTNA+SG+ +G + G + T LA++
Sbjct 501 FGLAGIVGYHTVWGVTPALHSPLMSVTNAISGLTAVGGLSLMGGGYLPSSTAETLAVLAA 560
Query 985 FLASVNVAGGFFVTHRMLKMFQ 1006
F++SVN+AGGF VT RML MF+
Sbjct 561 FISSVNIAGGFLVTQRMLDMFK 582
> hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide
transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 370 bits (949), Expect = 2e-101, Method: Compositional matrix adjust.
Identities = 200/402 (49%), Positives = 263/402 (65%), Gaps = 7/402 (1%)
Query 54 LGLYIAQSVNMTEMPQLVALFHSFVGLAAVMVGFANF--HSPAGVERASSLLRLLEVYAG 111
+GL IA+ + ++++PQLVA FHS VGLAAV+ A + P A++ L + Y G
Sbjct 686 IGLTIAKRIQISDLPQLVAAFHSLVGLAAVLTCIAEYIIEYPHFATDAAANLTKIVAYLG 745
Query 112 VFVAGITFTGSVVAAAKLHGSMESRSLRVPGRHALNTATIAAIGVLGAL-FCVSSGHFTR 170
++ G+TF+GS++A KL G ++S L +PGRH LN +AA V G + F V T
Sbjct 746 TYIGGVTFSGSLIAYGKLQGLLKSAPLLLPGRHLLNAGLLAA-SVGGIIPFMVDPSFTTG 804
Query 171 MLCLYVNAGLSMWLGFHLVAAIGGADMPVVISLLNSYSGVALAASGFMLDNNLLIIAGAL 230
+ CL + LS +G L AAIGGADMPVVI++LNSYSG AL A GF+L+NNLL I GAL
Sbjct 805 ITCLGSVSALSAVMGVTLTAAIGGADMPVVITVLNSYSGWALCAEGFLLNNNLLTIVGAL 864
Query 231 IASSGAILSYIMCKGMNRSLWNVVLGGFEEAEEVGAASPQ--GAVQQATADQVADELLAA 288
I SSGAILSYIMC MNRSL NV+LGG+ G + G + D D + A
Sbjct 865 IGSSGAILSYIMCVAMNRSLANVILGGYGTTSTAGGKPMEISGTHTEINLDNAIDMIREA 924
Query 289 RKVLIVPGYGMAVARCQSELADIAKNLMNCGITVDFGIHPVAGRMPGHMNVLLAEADVPY 348
++I PGYG+ A+ Q +AD+ K L G V FGIHPVAGRMPG +NVLLAEA VPY
Sbjct 925 NSIIITPGYGLCAAKAQYPIADLVKMLTEQGKKVRFGIHPVAGRMPGQLNVLLAEAGVPY 984
Query 349 KIVKEMSEVNPEMSSYDVVLVVGANDTVNPAALE-PGSKISGMPVIEAWKARRVFVLKRS 407
IV EM E+N + D+VLV+GANDTVN AA E P S I+GMPV+E WK+++V V+KRS
Sbjct 985 DIVLEMDEINHDFPDTDLVLVIGANDTVNSAAQEDPNSIIAGMPVLEVWKSKQVIVMKRS 1044
Query 408 MAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARVNARAEQ 449
+ GYA+++NP+F+ NT ML G+AK T A+ A+V ++
Sbjct 1045 LGVGYAAVDNPIFYKPNTAMLLGDAKKTCDALQAKVRESYQK 1086
Score = 313 bits (802), Expect = 2e-84, Method: Compositional matrix adjust.
Identities = 204/556 (36%), Positives = 309/556 (55%), Gaps = 49/556 (8%)
Query 478 PYPRMAVGVLRD-SNGSVMVPVAPKFVPKLRKLAFRVNVESGAGADAGFTDEEYRRAGAE 536
PY ++ VGV ++ V ++P V L K F V VESGAG + F+D+ YR AGA+
Sbjct 53 PYKQLTVGVPKEIFQNEKRVALSPAGVQNLVKQGFNVVVESGAGEASKFSDDHYRVAGAQ 112
Query 537 VLSGPDAVINQSQVLLRVSAP--SPDL----VSRIPRDKVLISYLFPSINQQALDMLARQ 590
+ + + S ++++V AP +P L + LIS+++P+ N + L+ L+++
Sbjct 113 IQGAKEVL--ASDLVVKVRAPMVNPTLGVHEADLLKTSGTLISFIYPAQNPELLNKLSQR 170
Query 591 GVTALAVDEVPRVTRAQKLDVKSAMQGLQGYRAVIEAFNALPKLSKASISAAGRVEAAKV 650
T LA+D+VPRVT AQ D S+M + GY+AV+ A N + I+AAG+V AK+
Sbjct 171 KTTVLAMDQVPRVTIAQGYDALSSMANIAGYKAVVLAANHFGRFFTGQITAAGKVPPAKI 230
Query 651 FVIGAGVAGLQAISTAHGLGAQVFGHDVRSATREEVESCGGKFIGLRMGEEGEVLGGYAR 710
++G GVAGL + A +GA V G D R+A E+ +S G + + + + E GE GGYA+
Sbjct 231 LIVGGGVAGLASAGAAKSMGAIVRGFDTRAAALEQFKSLGAEPLEVDLKESGEGQGGYAK 290
Query 711 EMGDAYQRAQREMIANTIKHCDVVICTAAIHGRPSPKLISRDMLRSMKPGSVVVDLATEF 770
EM + A+ ++ A K D++I TA I G+ +P L +++M+ SMK GSVVVDLA E
Sbjct 291 EMSKEFIEAEMKLFAQQCKEVDILISTALIPGKKAPVLFNKEMIESMKEGSVVVDLAAE- 349
Query 771 GDVRSGWGGNVEVSPKDDQIVVDGVTVIGRRRIETRMPIQASELFSMNICNLLEDLG-GG 829
GGN E + + + G+T IG + +RM QAS L+S NI LL+ +
Sbjct 350 ------AGGNFETTKPGELYIHKGITHIGYTDLPSRMATQASTLYSNNITKLLKAISPDK 403
Query 830 SNFRINMDDE--------VIRGLVAVYQGRNVWQPSQPTPVSRTPPRGQMPPPSAPGAPA 881
NF ++ D+ VIRG V + G+ ++ P P + P+ GAP
Sbjct 404 DNFYFDVKDDFDFGTMGHVIRGTVVMKDGKVIF----PAPTPKNIPQ---------GAPV 450
Query 882 PEKPGA--FAQALASDAFFAMCLVVAAA----VVGLLGIVLDPVEL---KHLTLLGLSLI 932
+K A A+ A+ F + A+A + G+LG+ + L + +T GL+ I
Sbjct 451 KQKTVAELEAEKAATITPFRKTMSTASAYTAGLTGILGLGIAAPNLAFSQMVTTFGLAGI 510
Query 933 VGYYCVWAVTPSLHTPLMSVTNALSGVIVIGCMLEYGTAMISGFTL--LALIGTFLASVN 990
VGY+ VW VTP+LH+PLMSVTNA+SG+ +G + G + T LA + F++SVN
Sbjct 511 VGYHTVWGVTPALHSPLMSVTNAISGLTAVGGLALMGGHLYPSTTSQGLAALAAFISSVN 570
Query 991 VAGGFFVTHRMLKMFQ 1006
+AGGF VT RML MF+
Sbjct 571 IAGGFLVTQRMLDMFK 586
> mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 369 bits (948), Expect = 2e-101, Method: Compositional matrix adjust.
Identities = 200/402 (49%), Positives = 262/402 (65%), Gaps = 7/402 (1%)
Query 54 LGLYIAQSVNMTEMPQLVALFHSFVGLAAVMVGFANF--HSPAGVERASSLLRLLEVYAG 111
+GL IA+ + ++++PQLVA FHS VGLAAV+ A + P A+S + Y G
Sbjct 686 IGLTIAKRIQISDLPQLVAAFHSLVGLAAVLTCMAEYIVEYPHFAMDATSNFTKIVAYLG 745
Query 112 VFVAGITFTGSVVAAAKLHGSMESRSLRVPGRHALNTATIAAIGVLGAL-FCVSSGHFTR 170
++ G+TF+GS+VA KL G ++S L +PGRHALN +AA V G + F T
Sbjct 746 TYIGGVTFSGSLVAYGKLQGILKSAPLLLPGRHALNAGLLAA-SVGGIIPFMADPSFTTG 804
Query 171 MLCLYVNAGLSMWLGFHLVAAIGGADMPVVISLLNSYSGVALAASGFMLDNNLLIIAGAL 230
+ CL + LS +G L AAIGGADMPVVI++LNSYSG AL A GF+L+NNLL I GAL
Sbjct 805 ITCLGSVSALSTLMGVTLTAAIGGADMPVVITVLNSYSGWALCAEGFLLNNNLLTIVGAL 864
Query 231 IASSGAILSYIMCKGMNRSLWNVVLGGFEEAEEVGAASPQ--GAVQQATADQVADELLAA 288
I SSGAILSYIMC MNRSL NV+LGG+ G + G + D + + A
Sbjct 865 IGSSGAILSYIMCVAMNRSLANVILGGYGTTSTAGGKPMEISGTHTEINLDNAVEMIREA 924
Query 289 RKVLIVPGYGMAVARCQSELADIAKNLMNCGITVDFGIHPVAGRMPGHMNVLLAEADVPY 348
++I PGYG+ A+ Q +AD+ K L G V FGIHPVAGRMPG +NVLLAEA VPY
Sbjct 925 NSIVITPGYGLCAAKAQYPIADLVKMLTEQGKKVRFGIHPVAGRMPGQLNVLLAEAGVPY 984
Query 349 KIVKEMSEVNPEMSSYDVVLVVGANDTVNPAALE-PGSKISGMPVIEAWKARRVFVLKRS 407
IV EM E+N + D+VLV+GANDTVN AA E P S I+GMPV+E WK+++V V+KRS
Sbjct 985 DIVLEMDEINSDFPDTDLVLVIGANDTVNSAAQEDPNSIIAGMPVLEVWKSKQVIVMKRS 1044
Query 408 MAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARVNARAEQ 449
+ GYA+++NP+F+ NT ML G+AK T A+ A+V ++
Sbjct 1045 LGVGYAAVDNPIFYKPNTAMLLGDAKKTCDALQAKVRESYQK 1086
Score = 313 bits (801), Expect = 3e-84, Method: Compositional matrix adjust.
Identities = 211/575 (36%), Positives = 306/575 (53%), Gaps = 46/575 (8%)
Query 462 GLLEFDREERVDPSSW---------PYPRMAVGVLRD-SNGSVMVPVAPKFVPKLRKLAF 511
G +F R R + W PY ++ VGV ++ V ++P V L K F
Sbjct 28 GKRDFVRMLRTHQALWCKSPVKPGIPYKQLTVGVPKEIFQNEKRVALSPAGVQALVKQGF 87
Query 512 RVNVESGAGADAGFTDEEYRRAGAEVLSGPDAVINQSQVLLRVSAP--SPDLVSR----I 565
V VESGAG + F D+ YR AGA++ G V+ S ++++V AP +P L + +
Sbjct 88 NVVVESGAGEASKFPDDLYRAAGAQI-QGMKEVL-ASDLVVKVRAPMVNPTLGAHEADFL 145
Query 566 PRDKVLISYLFPSINQQALDMLARQGVTALAVDEVPRVTRAQKLDVKSAMQGLQGYRAVI 625
LIS+++P+ N L+ L+ + T LA+D+VPRVT AQ D S+M + GY+AV+
Sbjct 146 KPSGTLISFIYPAQNPDLLNKLSERKTTVLAMDQVPRVTIAQGYDALSSMANISGYKAVV 205
Query 626 EAFNALPKLSKASISAAGRVEAAKVFVIGAGVAGLQAISTAHGLGAQVFGHDVRSATREE 685
A N + I+AAG+V AK+ ++G GVAGL + A +GA V G D R+A E+
Sbjct 206 LAANHFGRFFTGQITAAGKVPPAKILIVGGGVAGLASAGAAKSMGAVVRGFDTRAAALEQ 265
Query 686 VESCGGKFIGLRMGEEGEVLGGYAREMGDAYQRAQREMIANTIKHCDVVICTAAIHGRPS 745
+S G + + + + E GE GGYA+EM + A+ ++ A K D++I TA I G+ +
Sbjct 266 FKSLGAEPLEVDLKESGEGQGGYAKEMSKEFIEAEMKLFAQQCKEVDILISTALIPGKKA 325
Query 746 PKLISRDMLRSMKPGSVVVDLATEFGDVRSGWGGNVEVSPKDDQIVVDGVTVIGRRRIET 805
P L S++M+ SMK GSVVVDLA E GGN E + + V G+T IG + +
Sbjct 326 PVLFSKEMIESMKEGSVVVDLAAE-------AGGNFETTKPGELYVHKGITHIGYTDLPS 378
Query 806 RMPIQASELFSMNICNLLEDLG-GGSNFRINMDDE--------VIRGLVAVYQGRNVWQP 856
RM QAS L+S NI LL+ + NF + D+ VIRG V + G+ ++
Sbjct 379 RMATQASTLYSNNITKLLKAISPDKDNFHFEVKDDFDFGTMSHVIRGTVVMKDGKVIF-- 436
Query 857 SQPTPVSRTPPRGQMPPPSAPGAPAPEKPGAFAQALASDAFFAMCLVVAAAVVGL--LGI 914
P P + P P EK G + + V +A + G+ LGI
Sbjct 437 --PAPTPKNIPEEAPVKPKTVAELEAEKAGTVSMYTKT---LTTASVYSAGLTGMLGLGI 491
Query 915 VLDPVELKHL-TLLGLSLIVGYYCVWAVTPSLHTPLMSVTNALSGVIVIGCMLEYGTAMI 973
V V + T GL+ I+GY+ VW VTP+LH+PLMSVTNA+SG+ +G + G
Sbjct 492 VAPNVAFSQMVTTFGLAGIIGYHTVWGVTPALHSPLMSVTNAISGLTAVGGLALMGGHFY 551
Query 974 SGFT--LLALIGTFLASVNVAGGFFVTHRMLKMFQ 1006
T LA + TF++SVN+AGGF VT RML MF+
Sbjct 552 PSTTSQSLAALATFISSVNIAGGFLVTQRMLDMFK 586
> xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydrogenase
(EC:1.6.1.2); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1086
Score = 365 bits (937), Expect = 5e-100, Method: Compositional matrix adjust.
Identities = 195/397 (49%), Positives = 263/397 (66%), Gaps = 9/397 (2%)
Query 54 LGLYIAQSVNMTEMPQLVALFHSFVGLAAVMVGFANF---HSPAGVERASSLLRLLEVYA 110
+GL IA+ + ++++PQLVA FHS VGLAAV+ A + + + A++L +++ Y
Sbjct 686 IGLTIAKRIQISDLPQLVAAFHSLVGLAAVLTCVAEYMIEYPHFATDPAANLTKIV-AYL 744
Query 111 GVFVAGITFTGSVVAAAKLHGSMESRSLRVPGRHALNTATIAAIGVLGAL-FCVSSGHFT 169
G ++ G+TF+GS+VA KL G + S L +PGRH LN + A V G + + + + T
Sbjct 745 GTYIGGVTFSGSLVAYGKLQGVLNSAPLLLPGRHMLNAGLLTA-SVGGIIPYMLDPSYTT 803
Query 170 RMLCLYVNAGLSMWLGFHLVAAIGGADMPVVISLLNSYSGVALAASGFMLDNNLLIIAGA 229
+ CL + LS +G L AAIGGADMPVVI++LNSYSG AL A GF+L+NNLL I GA
Sbjct 804 GITCLGSVSALSAVMGVTLTAAIGGADMPVVITVLNSYSGWALCAEGFLLNNNLLTIVGA 863
Query 230 LIASSGAILSYIMCKGMNRSLWNVVLGGFEEAEEVGAASPQ--GAVQQATADQVADELLA 287
LI SSGAILSYIMC MNRSL NV+LGG+ G + G + D + +
Sbjct 864 LIGSSGAILSYIMCVAMNRSLTNVILGGYGTTSTAGGKPMEITGTHTEINLDNAVEYIRE 923
Query 288 ARKVLIVPGYGMAVARCQSELADIAKNLMNCGITVDFGIHPVAGRMPGHMNVLLAEADVP 347
A ++I PGYG+ A+ Q +AD+ K L G V FGIHPVAGRMPG +NVLLAEA VP
Sbjct 924 ANNIIITPGYGLCAAKAQYPIADLVKILKEAGKNVRFGIHPVAGRMPGQLNVLLAEAGVP 983
Query 348 YKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAALE-PGSKISGMPVIEAWKARRVFVLKR 406
Y IV EM E+N + D+VLV+GANDTVN AA E P S I+GMPV+E WK+++V V+KR
Sbjct 984 YDIVLEMDEINEDFPETDLVLVIGANDTVNSAAQEDPNSIIAGMPVLEVWKSKQVIVMKR 1043
Query 407 SMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARV 443
S+ GYA+++NP+F+ NT ML G+AK T ++ A+V
Sbjct 1044 SLGVGYAAVDNPIFYKPNTSMLLGDAKKTCDSLQAKV 1080
Score = 297 bits (760), Expect = 2e-79, Method: Compositional matrix adjust.
Identities = 211/558 (37%), Positives = 301/558 (53%), Gaps = 53/558 (9%)
Query 478 PYPRMAVGVLRD-SNGSVMVPVAPKFVPKLRKLAFRVNVESGAGADAGFTDEEYRRAGAE 536
PY ++ VGV ++ V ++P V L K F V VE+GAG + F+D+ Y+ AGA+
Sbjct 53 PYKQITVGVPKEIFQNEKRVALSPAGVQALVKQGFNVVVETGAGEASKFSDDHYKEAGAK 112
Query 537 VLSGPDAVINQSQVLLRVSAP--SPDL----VSRIPRDKVLISYLFPSINQQALDMLARQ 590
+ D + S ++L+V AP +P L L+S+++P+ N L L+ +
Sbjct 113 IQGTKDVL--ASDLVLKVRAPMLNPALGVHEADMFKPSSTLVSFVYPAQNPDLLSKLSEK 170
Query 591 GVTALAVDEVPRVTRAQKLDVKSAMQGLQGYRAVIEAFNALPKLSKASISAAGRVEAAKV 650
+T LA+D+VPRVT AQ D S+M + GY+AV+ A N + I+AAG+V AKV
Sbjct 171 NMTVLAMDQVPRVTIAQGYDALSSMANISGYKAVVMAANNFGRFFTGQITAAGKVPPAKV 230
Query 651 FVIGAGVAGLQAISTAHGLGAQVFGHDVRSATREEVESCGGKFIGLRMGEEGEVLGGYAR 710
+IG GVAGL A A +GA V G D R+A E+ +S G + + + + E GE GGYA+
Sbjct 231 LIIGGGVAGLAAAGAAKSMGAIVRGFDTRAAALEQFKSLGAEPLEVDLKESGEGQGGYAK 290
Query 711 EMGDAYQRAQREMIANTIKHCDVVICTAAIHGRPSPKLISRDMLRSMKPGSVVVDLATEF 770
EM + A+ ++ A + D+++ TA I G+ +P L +DM+ MK GSVVVDLA E
Sbjct 291 EMSKEFIEAEMKLFAKQCQDVDIIVTTALIPGKTAPILFRKDMIELMKEGSVVVDLAAE- 349
Query 771 GDVRSGWGGNVEVSPKDDQIVVDGVTVIGRRRIETRMPIQASELFSMNICNLLEDLG-GG 829
GGN+E + D V GVT IG I +RM QAS L+S NI LL+ +
Sbjct 350 ------AGGNIETTKPGDIYVHKGVTHIGYTDIPSRMASQASTLYSNNITKLLKAISPDK 403
Query 830 SNFRINMDDE--------VIRGLVAVYQGRNVWQPSQPTPVSRTPPRGQMPPPSAPGAPA 881
NF ++ D+ VIRG V + G NV P+ PPP+ A
Sbjct 404 DNFYYDIKDDFDYGTMDHVIRGTVVMKDG-NVIFPA--------------PPPNKIPQAA 448
Query 882 PEKPGAFAQALASDAF----FAMCLVVAAAVVGLLGIVLD-------PVELKHLTLLGLS 930
P K + AQ A A F + AAA LG +L + +T GL+
Sbjct 449 PVKQKSVAQIEAEKAASISPFRKTMNGAAAYTAGLGTLLSLGIASPHSAFTQMVTTFGLA 508
Query 931 LIVGYYCVWAVTPSLHTPLMSVTNALSGVIVIGCMLEYGTAMISGFT--LLALIGTFLAS 988
IVGY+ VW VTP+LH+PLMSVTNA+SG+ +G + G + T LLA++ F++S
Sbjct 509 GIVGYHTVWGVTPALHSPLMSVTNAISGLTAVGGLALMGGGYLPTNTHELLAVLAAFVSS 568
Query 989 VNVAGGFFVTHRMLKMFQ 1006
+N+AGGF VT RML MF+
Sbjct 569 INIAGGFLVTQRMLDMFK 586
> cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase
family member (nnt-1); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1041
Score = 361 bits (927), Expect = 8e-99, Method: Compositional matrix adjust.
Identities = 194/400 (48%), Positives = 262/400 (65%), Gaps = 8/400 (2%)
Query 54 LGLYIAQSVNMTEMPQLVALFHSFVGLAAVMVGFANF--HSPAGVERASSLLRLLEVYA- 110
+GL IA + +T++PQLVA FHSFVGLAA + ANF P +E S+
Sbjct 641 IGLGIANRIKVTDLPQLVAAFHSFVGLAATLTCLANFIQEHPHFLEDPSNAAAAKLALFL 700
Query 111 GVFVAGITFTGSVVAAAKLHGSMESRSLRVPGRHALNTATIAA-IGVLGALFCVSSGHFT 169
G ++ G+TFTGS++A KL G + S +P RH LN A +A +G LG + S+ T
Sbjct 701 GTYIGGVTFTGSLMAYGKLQGILASAPTYLPARHVLNGALLAGNVGALGT-YMYSTDFGT 759
Query 170 RMLCLYVNAGLSMWLGFHLVAAIGGADMPVVISLLNSYSGVALAASGFMLDNNLLIIAGA 229
M L GLS +G L AIGGADMP+VI++LNSYSG AL A GFMLDN+LL + GA
Sbjct 760 GMSMLGGTVGLSSLMGVTLTMAIGGADMPIVITVLNSYSGWALCAEGFMLDNSLLTVLGA 819
Query 230 LIASSGAILSYIMCKGMNRSLWNVVLGGF-EEAEEVGAASP-QGAVQQATADQVADELLA 287
LI SSGAILS+IMCK MNRSL NV+LGG +++ G A +G ++ + AD LL
Sbjct 820 LIGSSGAILSHIMCKAMNRSLLNVILGGVGTKSKGTGEAKAIEGTAKEIAPVETADMLLN 879
Query 288 ARKVLIVPGYGMAVARCQSELADIAKNLMNCGITVDFGIHPVAGRMPGHMNVLLAEADVP 347
AR V+I+PGYG+ A+ Q +A + K L G+ V F IHPVAGRMPG +NVLLAEA VP
Sbjct 880 ARSVIIIPGYGLCAAQAQYPIAQLVKELQQRGVRVRFAIHPVAGRMPGQLNVLLAEAGVP 939
Query 348 YKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAAL-EPGSKISGMPVIEAWKARRVFVLKR 406
Y IV+EM E+N + DV LV+G+NDT+N AA +P S I+GMPV+ W +++V ++KR
Sbjct 940 YDIVEEMEEINEDFKETDVALVIGSNDTINSAAEDDPNSSIAGMPVLRVWNSKQVIIVKR 999
Query 407 SMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARVNAR 446
++ GYA+++NP+F ENT+ML G+AK + + V ++
Sbjct 1000 TLGTGYAAVDNPVFFNENTQMLLGDAKKMSEKLLEEVKSK 1039
Score = 352 bits (903), Expect = 4e-96, Method: Compositional matrix adjust.
Identities = 214/532 (40%), Positives = 300/532 (56%), Gaps = 21/532 (3%)
Query 479 YPRMAVGVLRD-SNGSVMVPVAPKFVPKLRKLAFRVNVESGAGADAGFTDEEYRRAGAEV 537
Y ++ V V ++ G V ++P V L+K V +E AG AG+++EEY R+GA+V
Sbjct 26 YSKLKVAVPKEIFPGEKRVSLSPNGVALLKKNGISVLIEENAGVLAGYSNEEYVRSGADV 85
Query 538 LSGPDAVINQSQVLLRVSAPSPDLVSRIPRDKVLISYLFPSINQQALDMLARQGVTALAV 597
G + + ++L+V P+ + VS++ LIS++ P NQ LD L + T A+
Sbjct 86 --GKHNEVFNTDIMLKVRPPTENEVSKLKSGCTLISFIHPGQNQALLDSLTKTDKTVFAM 143
Query 598 DEVPRVTRAQKLDVKSAMQGLQGYRAVIEAFNALPKLSKASISAAGRVEAAKVFVIGAGV 657
D VPR++RAQ D S+M + GYRAVIEA N + I+AAG+V AKV VIG GV
Sbjct 144 DCVPRISRAQVFDALSSMANIAGYRAVIEAANHFGRFFTGQITAAGKVPPAKVLVIGGGV 203
Query 658 AGLQAISTAHGLGAQVFGHDVRSATREEVESCGGKFIGLRMGEEGEVLGGYAREMGDAYQ 717
AGL AI T+ G+GA V G D R+A +E VES G +F+ + + E+GE GGYA+EM +
Sbjct 204 AGLSAIGTSRGMGAVVRGFDTRAAVKEHVESLGAQFLTVNVKEDGEGGGGYAKEMSKEFI 263
Query 718 RAQREMIANTIKHCDVVICTAAIHGRPSPKLISRDMLRSMKPGSVVVDLATEFGDVRSGW 777
A+ ++ A+ K D++I TA I G+ +P LI+ +M++SMKPGSVVVDLA E
Sbjct 264 DAEMKLFADQCKDVDIIITTALIPGKKAPILITEEMIKSMKPGSVVVDLAAE-------S 316
Query 778 GGNVEVSPKDDQIVVDGVTVIGRRRIETRMPIQASELFSMNICNLLEDLGGGSNFRINMD 837
GGN+ + + V GVT IG + +R+P Q+SEL+S NI L LG F +N +
Sbjct 317 GGNIATTRPGEVYVKHGVTHIGFTDLPSRLPTQSSELYSNNIAKFLLHLGKDKTFFVNEE 376
Query 838 DEVIRGLVAVYQGRNVWQPSQPTPVSRTPPRGQMPPPSAPGAPAPEKPGAFAQALASDAF 897
DEV RG + V G+ W P P+ PS A P P A
Sbjct 377 DEVARGALVVRDGQMKWPPPPINFPPPAAPKSD--KPSENTALVPLTP------FRKTAN 428
Query 898 FAMCLVVAAAVVGLLGIV-LDPVELKHLTLLGLSLIVGYYCVWAVTPSLHTPLMSVTNAL 956
+ L V LLGI +P T L+ +VGY+ VW VTP+LH+PLMSVTNA+
Sbjct 429 QTLLLTSGLGSVSLLGIAGTNPQISSMSTTFALAGLVGYHTVWGVTPALHSPLMSVTNAI 488
Query 957 SGVIVIG--CMLEYGTAMISGFTLLALIGTFLASVNVAGGFFVTHRMLKMFQ 1006
SG G C++ G + +AL+ TF++SVN+ GGF VT RML MF+
Sbjct 489 SGTTAAGALCLMGGGLMPQNSAQTMALLATFISSVNIGGGFLVTKRMLDMFK 540
> eco:b1603 pntA, ECK1598, JW1595; pyridine nucleotide transhydrogenase,
alpha subunit (EC:1.6.1.2); K00324 NAD(P) transhydrogenase
subunit alpha [EC:1.6.1.2]
Length=510
Score = 350 bits (898), Expect = 2e-95, Method: Compositional matrix adjust.
Identities = 208/529 (39%), Positives = 303/529 (57%), Gaps = 25/529 (4%)
Query 482 MAVGVLRDS-NGSVMVPVAPKFVPKLRKLAFRVNVESGAGADAGFTDEEYRRAGAEVLSG 540
M +G+ R+ V PK V +L KL F V VESGAG A F D+ + +AGAE++ G
Sbjct 1 MRIGIPRERLTNETRVAATPKTVEQLLKLGFTVAVESGAGQLASFDDKAFVQAGAEIVEG 60
Query 541 PDAVINQSQVLLRVSAPSPDLVSRIPRDKVLISYLFPSINQQALDMLARQGVTALAVDEV 600
+ QS+++L+V+AP D ++ + L+S+++P+ N + + LA + VT +A+D V
Sbjct 61 NS--VWQSEIILKVNAPLDDEIALLNPGTTLVSFIWPAQNPELMQKLAERNVTVMAMDSV 118
Query 601 PRVTRAQKLDVKSAMQGLQGYRAVIEAFNALPKLSKASISAAGRVEAAKVFVIGAGVAGL 660
PR++RAQ LD S+M + GYRA++EA + + I+AAG+V AKV VIGAGVAGL
Sbjct 119 PRISRAQSLDALSSMANIAGYRAIVEAAHEFGRFFTGQITAAGKVPPAKVMVIGAGVAGL 178
Query 661 QAISTAHGLGAQVFGHDVRSATREEVESCGGKFIGLRMGEEGEVLGGYAREMGDAYQRAQ 720
AI A+ LGA V D R +E+V+S G +F+ L EE GYA+ M DA+ +A+
Sbjct 179 AAIGAANSLGAIVRAFDTRPEVKEQVQSMGAEFLELDFKEEAGSGDGYAKVMSDAFIKAE 238
Query 721 REMIANTIKHCDVVICTAAIHGRPSPKLISRDMLRSMKPGSVVVDLATEFGDVRSGWGGN 780
E+ A K D+++ TA I G+P+PKLI+R+M+ SMK GSV+VDLA + GGN
Sbjct 239 MELFAAQAKEVDIIVTTALIPGKPAPKLITREMVDSMKAGSVIVDLAAQN-------GGN 291
Query 781 VEVS-PKDDQIVVDGVTVIGRRRIETRMPIQASELFSMNICNLLEDL--GGGSNFRINMD 837
E + P + +GV VIG + R+P Q+S+L+ N+ NLL+ L N ++ D
Sbjct 292 CEYTVPGEIFTTENGVKVIGYTDLPGRLPTQSSQLYGTNLVNLLKLLCKEKDGNITVDFD 351
Query 838 DEVIRGLVAVYQGRNVWQPSQPTPVSRTPPRGQMPPPSAPGAPAPEKPGAFAQALASDAF 897
D VIRG+ + G W P+ P VS + A
Sbjct 352 DVVIRGVTVIRAGEITW-PAPPIQVSAQ-----PQAAQKAAPEVKTEEKCTCSPWRKYAL 405
Query 898 FAMCLVVAAAVVGLLGIVLDPVELKHLTLLGLSLIVGYYCVWAVTPSLHTPLMSVTNALS 957
A+ ++ + G + V L H T+ L+ +VGYY VW V+ +LHTPLMSVTNA+S
Sbjct 406 MALAII----LFGWMASVAPKEFLGHFTVFALACVVGYYVVWNVSHALHTPLMSVTNAIS 461
Query 958 GVIVIGCMLEYGTAMISGFTLLALIGTFLASVNVAGGFFVTHRMLKMFQ 1006
G+IV+G +L+ G F L+ I +AS+N+ GGF VT RMLKMF+
Sbjct 462 GIIVVGALLQIGQGGWVSF--LSFIAVLIASINIFGGFTVTQRMLKMFR 508
> dre:100331743 Nicotinamide Nucleotide Transhydrogenase family
member (nnt-1)-like
Length=518
Score = 139 bits (350), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 107/298 (35%), Positives = 158/298 (53%), Gaps = 28/298 (9%)
Query 723 MIANTIKHCDVVICTAAIHGRPSPKLISRDMLRSMKPGSVVVDLATEFGDVRSGWGGNVE 782
+ A + D++I TA G+ +P LI R+M+ SM+ GSVVVDLA E GGN+E
Sbjct 3 LFAKQCRDVDIIISTALNPGKRAPVLIKREMVESMRDGSVVVDLAAE-------AGGNIE 55
Query 783 VSPKDDQIVVDGVTVIGRRRIETRMPIQASELFSMNICNLLEDLGGGSNFRINMD----- 837
+ + V GVT IG + +RM QAS L+S NI LL+ + F N +
Sbjct 56 TTKPGELYVHQGVTHIGYTDLPSRMATQASSLYSNNIIKLLKAISPDKEF-FNFEPTDEF 114
Query 838 -----DEVIRGLVAVYQGRNVWQPSQPTPVSRTPPRGQMPPPSAPGAPAPEKP--GAFAQ 890
D VIRG + + QG+N++ P PP Q S A ++ F +
Sbjct 115 DYGTLDHVIRGTMVMKQGQNLFPAPLPKTTPPPPPAKQ---KSVAELEAEKRAVISPFRR 171
Query 891 ALASDAFFAMCLVVAAAVVGLLGIVLDPVELKHLTLLGLSLIVGYYCVWAVTPSLHTPLM 950
+ S + L + V+GL + + +T GL+ IVGY+ VW VTP+LH+PLM
Sbjct 172 TMTSAGVYTAGL---STVLGLGIASPNAAFTQMVTTFGLAGIVGYHTVWGVTPALHSPLM 228
Query 951 SVTNALSGVIVIGCMLEYGTAMI--SGFTLLALIGTFLASVNVAGGFFVTHRMLKMFQ 1006
SVTNA+SG+ +G ++ G + S LAL+ F++S+N+AGGF +T +ML MF+
Sbjct 229 SVTNAISGLTAVGGLVLMGGGLTPSSLPESLALLAAFVSSINIAGGFLITQKMLDMFK 286
Score = 86.7 bits (213), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 54/132 (40%), Positives = 82/132 (62%), Gaps = 6/132 (4%)
Query 54 LGLYIAQSVNMTEMPQLVALFHSFVGLAAVMVGFANF---HSPAGVERASSLLRLLEVYA 110
LGL IA+ + ++++PQLVA FHS VGLAAV+ A + + V A+ +L+++ Y
Sbjct 386 LGLTIAKRIEISDLPQLVAAFHSLVGLAAVLTCVAEYMIEYPHLDVHPAAGVLKIV-AYL 444
Query 111 GVFVAGITFTGSVVAAAKLHGSMESRSLRVPGRHALNTA-TIAAIGVLGALFCVSSGHFT 169
G ++ G+TF+GS+VA KL G ++S L +PGRH LN +A++G + F +S T
Sbjct 445 GTYIGGVTFSGSLVAYGKLQGLLDSAPLLLPGRHMLNAGLMLASVGGM-VPFMLSDSFHT 503
Query 170 RMLCLYVNAGLS 181
M CL +GLS
Sbjct 504 GMGCLLGVSGLS 515
> tgo:TGME49_003500 alanine dehydrogenase, putative (EC:1.4.1.1)
Length=462
Score = 78.2 bits (191), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 80/301 (26%), Positives = 133/301 (44%), Gaps = 29/301 (9%)
Query 496 VPVAPKFVPKLRKLAFRVNVESGAGADAGFTDEEYRRAGAEVLSGPDAVINQSQVLLRVS 555
V + P V +L +V V++GAG +GFTDE Y AGA ++ + V SQ++++V
Sbjct 43 VALTPATVAELVNNGHKVVVQTGAGVASGFTDESYTAAGATMVQTTEEVYKTSQMIVKVQ 102
Query 556 APSPDLVSRIPRDKVLISYLFPSINQQALDMLARQGVTALAVDEVPRVTRAQKLDVKSAM 615
AP P S I +V+ ++ N+ + + ++ + +++ T + AM
Sbjct 103 APQPQEYSFIQPGQVIFAFFSFENNKTLFETMMQREAVCFSYEQMK--TETGTTPILQAM 160
Query 616 QGLQGYRAVIEAFNALPKLSKASISAAGRV---EAAKVFVIGAGVAGLQAISTAHGLGAQ 672
+ G A+ +A L K S G V + V V+G GVA +QA A GAQ
Sbjct 161 CEISGQLAISQAMKYLEKTMGGSGCMLGTVTGMTSGYVLVLGGGVAAMQAARVAASTGAQ 220
Query 673 VFGHDV-----RSATREEVESCGGKFIG----LRMGEEGEVLGGYAREMGDAYQRAQREM 723
V DV R+ T ++C + ++ ++ +V+ +G + Q M
Sbjct 221 VCIMDVCMSNMRTMTEMLPKNCTTMYFCTENLVQQIQKADVV------IGAVFGSFQ--M 272
Query 724 IANTIKHCDVVICTAAIHGRPSPKLISRDMLRSMKPGSVVVDLATEFGDVRSGWGGNVEV 783
A + T +P L+ ++ + MKPGSV+VDLA GGN E
Sbjct 273 PATVTNTTVMNQKTTMTQVTKAPMLVKKEHVAMMKPGSVIVDLAV-------SRGGNFET 325
Query 784 S 784
+
Sbjct 326 T 326
> tgo:TGME49_115260 alanine dehydrogenase, putative (EC:1.4.1.1)
Length=390
Score = 55.5 bits (132), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 32/122 (26%), Positives = 60/122 (49%), Gaps = 0/122 (0%)
Query 496 VPVAPKFVPKLRKLAFRVNVESGAGADAGFTDEEYRRAGAEVLSGPDAVINQSQVLLRVS 555
V +AP +L K V +E AG A FTDE+Y + GAE+++ + + +S+++++V
Sbjct 16 VGLAPAGALELVKAGHTVLIEKDAGKKAHFTDEDYVQQGAEIVASAEELYGRSEMIVKVK 75
Query 556 APSPDLVSRIPRDKVLISYLFPSINQQALDMLARQGVTALAVDEVPRVTRAQKLDVKSAM 615
P PD + ++L Y +Q + + G ++ + V + R L+ S +
Sbjct 76 EPQPDEWKLVKSGQILFCYFHFCASQSLTEAMLNSGAICISYETVQKDGRLPLLEPMSEV 135
Query 616 QG 617
G
Sbjct 136 AG 137
Score = 46.2 bits (108), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 25/70 (35%), Positives = 40/70 (57%), Gaps = 5/70 (7%)
Query 731 CDVVICTAAIHGRPSPKLISRDMLRSMKPGSVVVDLATEFGDVRSGWGGNVEVSPKDDQI 790
CD+++ + GR +PK++ R+ L+ MKPGSV+VD++ + G G V+ D I
Sbjct 231 CDLLVGAVLLPGRKAPKIVRREHLKRMKPGSVIVDVSIDHG----GCVETTRVTCHHDPI 286
Query 791 V-VDGVTVIG 799
VDG+ G
Sbjct 287 YEVDGIIHYG 296
> dre:100331681 MGC83563 protein-like
Length=142
Score = 48.1 bits (113), Expect = 2e-04, Method: Composition-based stats.
Identities = 29/60 (48%), Positives = 38/60 (63%), Gaps = 2/60 (3%)
Query 496 VPVAPKFVPKLRKLAFRVNVESGAGADAGFTDEEYRRAGAEVLSGPDAVINQSQVLLRVS 555
V V+P V L K F V VESGAGA A F+DE+YR AGA++ A+ S ++L+VS
Sbjct 56 VSVSPAGVELLVKQGFSVCVESGAGAAAQFSDEQYRAAGAKITDTHTAL--ASHLVLKVS 113
> dre:556058 si:ch211-194c3.5
Length=1157
Score = 36.2 bits (82), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 34/75 (45%), Gaps = 0/75 (0%)
Query 223 LLIIAGALIASSGAILSYIMCKGMNRSLWNVVLGGFEEAEEVGAASPQGAVQQATADQVA 282
L IIAG + + ++M N S W V E + E A+ Q + A Q
Sbjct 748 LTIIAGLPGSHKENLCDFLMEVNQNSSRWEVFCPALEGSGEFSASHLQRFLSSLLAKQRE 807
Query 283 DELLAARKVLIVPGY 297
++ + R VL++PGY
Sbjct 808 TDMNSFRVVLLIPGY 822
> dre:321756 tgif1, Tgif, fa06h05, fb33g11, wu:fa06h05, wu:fb33g11,
zgc:55852; TGFB-induced factor homeobox 1
Length=273
Score = 36.2 bits (82), Expect = 0.79, Method: Composition-based stats.
Identities = 17/47 (36%), Positives = 22/47 (46%), Gaps = 4/47 (8%)
Query 843 GLVAVYQGRNVWQPSQPTPVSRTPPRGQMP----PPSAPGAPAPEKP 885
GL+A + RN ++P P P S TP P P P +P P P
Sbjct 132 GLLANGEDRNSYEPGSPHPTSNTPTSNGYPKKALSPKTPPSPGPVLP 178
> hsa:3664 IRF6, LPS, OFC6, PIT, PPS, VWS, VWS1; interferon regulatory
factor 6; K10154 interferon regulatory factor 6
Length=467
Score = 34.3 bits (77), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 32/145 (22%), Positives = 60/145 (41%), Gaps = 14/145 (9%)
Query 736 CTAAIHGRPSPKLISRDMLRSMKPGSVVVDLATEFGDVRSGWGGNVEVSPKDDQIVVDGV 795
C G +P L++ +++ K + L T D+ + G +E P + + G
Sbjct 319 CKVYWSGPCAPSLVAPNLIERQKKVKLFC-LETFLSDLIAHQKGQIEKQPPFEIYLCFGE 377
Query 796 TVIGRRRIETRM------PIQASELFSMNICNLLEDLGGGS-NFRI---NMDDEVIRGLV 845
+ +E ++ P+ A ++ M + GS +I ++ D ++ L
Sbjct 378 EWPDGKPLERKLILVQVIPVVARMIYEMFSGDFTRSFDSGSVRLQISTPDIKDNIVAQLK 437
Query 846 AVY---QGRNVWQPSQPTPVSRTPP 867
+Y Q + WQP QPTP + PP
Sbjct 438 QLYRILQTQESWQPMQPTPSMQLPP 462
> mmu:654821 Gcnt7, A330041C17Rik; glucosaminyl (N-acetyl) transferase
family member 7 (EC:2.4.1.-)
Length=433
Score = 33.9 bits (76), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 27/108 (25%), Positives = 46/108 (42%), Gaps = 25/108 (23%)
Query 779 GNVEVSPKDDQIVVDGVTVIGRRRIETRM---------PIQASELFSMNICNLLEDLGGG 829
GN+ +S K ++ D + RR++ + P Q + MN+C G
Sbjct 166 GNIFLSSKTQKVAHDNL-----RRLQAEIDCMRDLVHSPFQWH--YVMNLC--------G 210
Query 830 SNFRINMDDEVIRGLVAVYQGRNVWQPSQPTPVSRTPPRGQMPPPSAP 877
F I + E+I + ++G+N+ P P + P GQ PP +P
Sbjct 211 QEFPIKTNKEIIYDIRTRWKGKNI-TPGVTPPANSKPKTGQGPPKPSP 257
> hsa:29844 TFPT, FB1, INO80F, amida; TCF3 (E2A) fusion partner
(in childhood Leukemia); K11670 TCF3 fusion partner
Length=253
Score = 33.1 bits (74), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 39/82 (47%), Gaps = 5/82 (6%)
Query 801 RRIETRMPIQASELFSMNICNLLEDLGGGSNFRINMDDEVIRGLVAVYQGRNVWQPSQPT 860
+RI R+ Q F M + + D S F I ++DE +G A G +P P
Sbjct 107 QRITRRL--QQERRFLMRVLDSYGDDYRASQFTIVLEDEGSQGTDAPTPGNAENEP--PE 162
Query 861 PVSRTPPRGQMPPPSAPGAPAP 882
+ +PPR + P P PG+PAP
Sbjct 163 KETLSPPR-RTPAPPEPGSPAP 183
> sce:YMR020W FMS1; Fms1p (EC:1.5.3.11); K13367 non-specific polyamine
oxidase [EC:1.5.3.17]
Length=508
Score = 33.1 bits (74), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 15/24 (62%), Positives = 18/24 (75%), Gaps = 0/24 (0%)
Query 649 KVFVIGAGVAGLQAISTAHGLGAQ 672
KV +IGAG+AGL+A ST H G Q
Sbjct 10 KVIIIGAGIAGLKAASTLHQNGIQ 33
> cel:K05F1.6 hypothetical protein
Length=856
Score = 33.1 bits (74), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 30/118 (25%), Positives = 50/118 (42%), Gaps = 25/118 (21%)
Query 899 AMCLVVAAAVVGLLGIVLDPVELKHLTLLGLSLIVGYY-----------------CVWAV 941
++C+V A + G+V+ L + G +L GY+ CVW +
Sbjct 571 SVCIVFMATALVYYGLVM---ALSDQSAPGRTLFTGYFHLNNGIAGAIEIPTLFACVWMM 627
Query 942 TPSLHTPLMSVTNALSGVIVIGCMLEYGTAMISGFTLLALIGTFLASVNVAGGFFVTH 999
LM +T SG+ +I ML +M+SG +LAL + + V G F + +
Sbjct 628 QLGRKKALM-LTLITSGLFIIVAML----SMVSGHYMLALAFMYFGKIAVQGAFNILY 680
> cel:Y71F9AL.18 pme-1; Poly(ADP-ribose) Metabolism Enzyme family
member (pme-1); K10798 poly [ADP-ribose] polymerase [EC:2.4.2.30]
Length=945
Score = 32.7 bits (73), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 30/66 (45%), Gaps = 7/66 (10%)
Query 819 ICNLLEDLGG----GSNFRINMDDEVIRGLVAVYQGRNVWQPSQPTPVSRTPPRGQMPPP 874
I +ED+ G NF+I E + L A R +P+ P S TPP + P
Sbjct 162 ISGTVEDIPGWADYEENFKIKAVGEYVEALAA---KRRSTEPATPASASPTPPEAETPVL 218
Query 875 SAPGAP 880
SA G+P
Sbjct 219 SAEGSP 224
> mmu:387512 Tas2r135, Tas2r35, mt2r38; taste receptor, type 2,
member 135; K08474 taste receptor type 2
Length=321
Score = 32.3 bits (72), Expect = 9.4, Method: Composition-based stats.
Identities = 15/62 (24%), Positives = 32/62 (51%), Gaps = 0/62 (0%)
Query 146 LNTATIAAIGVLGALFCVSSGHFTRMLCLYVNAGLSMWLGFHLVAAIGGADMPVVISLLN 205
L ATI +LG +CV T + +++ L W+ + L++A+G + + ++ +
Sbjct 112 LTAATIWLCSLLGFFYCVKIATLTHPVFVWLKYRLPGWVPWMLLSAVGMSSLTSILCFIG 171
Query 206 SY 207
+Y
Sbjct 172 NY 173
Lambda K H
0.321 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 54665127920
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40