bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1197_orf3
Length=176
Score E
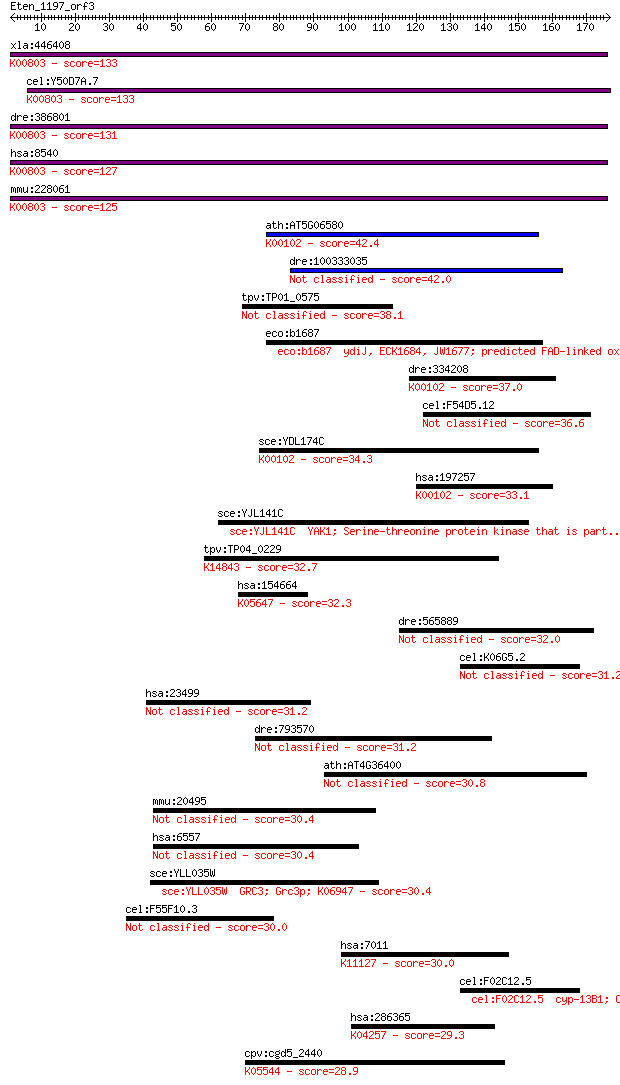
Sequences producing significant alignments: (Bits) Value
xla:446408 agps, MGC83829; alkylglycerone phosphate synthase (... 133 2e-31
cel:Y50D7A.7 ads-1; Alkyl-Dihydroxyacetonephosphate Synthase f... 133 3e-31
dre:386801 agps, fd16e06, wu:fd16e06, zgc:56718; alkylglyceron... 131 1e-30
hsa:8540 AGPS, ADAP-S, ADAS, ADHAPS, ADPS, ALDHPSY, DKFZp762O2... 127 2e-29
mmu:228061 Agps, 5832437L22, 9930035G10Rik, AW123847, Adaps, A... 125 8e-29
ath:AT5G06580 FAD linked oxidase family protein (EC:1.1.2.4); ... 42.4 9e-04
dre:100333035 lactate dehydrogenase D-like 42.0 0.001
tpv:TP01_0575 hypothetical protein 38.1 0.017
eco:b1687 ydiJ, ECK1684, JW1677; predicted FAD-linked oxidored... 37.7 0.020
dre:334208 ldhd, wu:fi36b04, zgc:55447; lactate dehydrogenase ... 37.0 0.037
cel:F54D5.12 hypothetical protein 36.6 0.049
sce:YDL174C DLD1; D-lactate dehydrogenase, oxidizes D-lactate ... 34.3 0.23
hsa:197257 LDHD, DLD, MGC57726; lactate dehydrogenase D (EC:1.... 33.1 0.50
sce:YJL141C YAK1; Serine-threonine protein kinase that is part... 33.1 0.55
tpv:TP04_0229 hypothetical protein; K14843 pescadillo 32.7 0.62
hsa:154664 ABCA13, DKFZp313D2411, FLJ16398, FLJ33876, FLJ33951... 32.3 0.87
dre:565889 d2hgdh, zgc:158661; D-2-hydroxyglutarate dehydrogen... 32.0 1.1
cel:K06G5.2 cyp-13B2; CYtochrome P450 family member (cyp-13B2) 31.2 2.0
hsa:23499 MACF1, ABP620, ACF7, FLJ45612, FLJ46776, KIAA0465, K... 31.2 2.1
dre:793570 btr07p; bloodthirsty-related gene family, member 7,... 31.2 2.1
ath:AT4G36400 FAD linked oxidase family protein 30.8 2.3
mmu:20495 Slc12a1, AI788571, D630042G03Rik, Nkcc2, mBSC1, ureh... 30.4 3.0
hsa:6557 SLC12A1, BSC1, MGC48843, NKCC2; solute carrier family... 30.4 3.1
sce:YLL035W GRC3; Grc3p; K06947 30.4 3.5
cel:F55F10.3 hypothetical protein 30.0 4.3
hsa:7011 TEP1, TLP1, TP1, TROVE1, VAULT2, p240; telomerase-ass... 30.0 4.3
cel:F02C12.5 cyp-13B1; CYtochrome P450 family member (cyp-13B1... 30.0 4.6
hsa:286365 OR13D1, OR9-15; olfactory receptor, family 13, subf... 29.3 6.3
cpv:cgd5_2440 Ylr401cp-like protein with 2 CCCH domains plus D... 28.9 8.8
> xla:446408 agps, MGC83829; alkylglycerone phosphate synthase
(EC:2.5.1.26); K00803 alkyldihydroxyacetonephosphate synthase
[EC:2.5.1.26]
Length=627
Score = 133 bits (335), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 66/178 (37%), Positives = 102/178 (57%), Gaps = 6/178 (3%)
Query 1 KVETCKWNGWGYCDTYVSMSSASMIRLHGSRYSLCGVDMGHWHEFMESIPGIEFAFSSPA 60
+ E KWNGWGY D+ + + G RY L G+ + E+ME G + +
Sbjct 56 RQEVLKWNGWGYNDSKFTFNKKGQAEFTGKRYKLSGMVLPALREWMEKTFGASLDQKTTS 115
Query 61 QDTLVI---PPQRVNNEAFIKELQQLAPEIEVANSEKERLFHGHGHTCNEIFELRYGTVA 117
+ +L + PP VN E F+++++ + I + ++R+F HGH +EIF LR G
Sbjct 116 RASLNVSDAPPALVN-EGFLQDIKAIG--ISYSQDTEDRIFRAHGHCLHEIFTLREGMFK 172
Query 118 RLPDVVLYPKEHKDVEAIVECASRHNVCLIPFGGGTSVTLGVAVPVSESRMVATVSLA 175
R+PD+V++P H+DV IV+ A +HNVCLIPFGGGTSV+ + P E R +A++ +
Sbjct 173 RIPDIVVWPSCHEDVVKIVDLACKHNVCLIPFGGGTSVSYALECPEDEKRTIASLDTS 230
> cel:Y50D7A.7 ads-1; Alkyl-Dihydroxyacetonephosphate Synthase
family member (ads-1); K00803 alkyldihydroxyacetonephosphate
synthase [EC:2.5.1.26]
Length=597
Score = 133 bits (335), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 67/174 (38%), Positives = 101/174 (58%), Gaps = 7/174 (4%)
Query 6 KWNGWGYCDTYVSMSSASMIRLHGSRYSLCGVDMGHWHEFMESIPGIEFAFSSPAQ---D 62
KWNGWGY D+ +++ + G +Y + G M H+ + E+ GI+ F SPAQ D
Sbjct 22 KWNGWGYSDSQFAINKDGHVTFTGDKYEISGKVMPHFRPWFENYLGIDLGFVSPAQKLSD 81
Query 63 TLVIPPQRVNNEAFIKELQQLAPEIEVANSEKERLFHGHGHTCNEIFELRYGTVARLPDV 122
++ P V NE I+ LQ+ +I +N + RL GHGHT +++ LR G + RLPD+
Sbjct 82 VIIDAP--VENEDIIEFLQE--NKISFSNEARIRLMRGHGHTVHDMINLREGKIPRLPDI 137
Query 123 VLYPKEHKDVEAIVECASRHNVCLIPFGGGTSVTLGVAVPVSESRMVATVSLAF 176
V++PK ++ I+E A HN +IP GGGTSVT + P +E R V ++ +A
Sbjct 138 VVWPKSEHEIVKIIEGAMSHNCAIIPIGGGTSVTNALDTPETEKRAVISMDMAL 191
> dre:386801 agps, fd16e06, wu:fd16e06, zgc:56718; alkylglycerone
phosphate synthase (EC:2.5.1.26); K00803 alkyldihydroxyacetonephosphate
synthase [EC:2.5.1.26]
Length=629
Score = 131 bits (330), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 68/179 (37%), Positives = 101/179 (56%), Gaps = 8/179 (4%)
Query 1 KVETCKWNGWGYCDTYVSMSSASMIRLHGSRYSLCGVDMGHWHEFMESIPGIEFAFSSPA 60
+ E KWNGWGY D+ + G RY L G+ + ++ E G SPA
Sbjct 58 RQEIMKWNGWGYSDSRFLFNKKGQAEFTGKRYRLSGLILPSLKDWFEGTFGASLQHKSPA 117
Query 61 QDTL----VIPPQRVNNEAFIKELQQLAPEIEVANSEKERLFHGHGHTCNEIFELRYGTV 116
+L V PP NEAF ++L+ A + ++ ++R+F HGH +EIF LR G +
Sbjct 118 TSSLNVSAVSPPNL--NEAFSEDLK--AAGLLASHDAEDRVFRAHGHCLHEIFALREGRI 173
Query 117 ARLPDVVLYPKEHKDVEAIVECASRHNVCLIPFGGGTSVTLGVAVPVSESRMVATVSLA 175
R+PD+V++P H DVE IV+ A +HNVCLIP+GGGTSV+ + P E+R + ++ +
Sbjct 174 GRVPDMVVWPSCHSDVEKIVDLACKHNVCLIPYGGGTSVSSALECPQEETRCIVSLDTS 232
> hsa:8540 AGPS, ADAP-S, ADAS, ADHAPS, ADPS, ALDHPSY, DKFZp762O2215,
FLJ99755; alkylglycerone phosphate synthase (EC:2.5.1.26);
K00803 alkyldihydroxyacetonephosphate synthase [EC:2.5.1.26]
Length=658
Score = 127 bits (320), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 63/177 (35%), Positives = 100/177 (56%), Gaps = 4/177 (2%)
Query 1 KVETCKWNGWGYCDTYVSMSSASMIRLHGSRYSLCGVDMGHWHEFMESIPGIEFAFSSPA 60
+ E KWNGWGY D+ + I L G RY L G+ + + E++++ G+ + +
Sbjct 87 RQEVMKWNGWGYNDSKFIFNKKGQIELTGKRYPLSGMGLPTFKEWIQNTLGVNVEHKTTS 146
Query 61 QDTL--VIPPQRVNNEAFIKELQQLAPEIEVANSEKERLFHGHGHTCNEIFELRYGTVAR 118
+ +L P V NE F+ +L++ I + +R+F HGH +EIF LR G R
Sbjct 147 KASLNPSDTPPSVVNEDFLHDLKE--TNISYSQEADDRVFRAHGHCLHEIFLLREGMFER 204
Query 119 LPDVVLYPKEHKDVEAIVECASRHNVCLIPFGGGTSVTLGVAVPVSESRMVATVSLA 175
+PD+VL+P H DV IV A ++N+C+IP GGGTSV+ G+ P E+R + ++ +
Sbjct 205 IPDIVLWPTCHDDVVKIVNLACKYNLCIIPIGGGTSVSYGLMCPADETRTIISLDTS 261
> mmu:228061 Agps, 5832437L22, 9930035G10Rik, AW123847, Adaps,
Adas, Adhaps, Adps, Aldhpsy, bs2; alkylglycerone phosphate
synthase (EC:2.5.1.26); K00803 alkyldihydroxyacetonephosphate
synthase [EC:2.5.1.26]
Length=671
Score = 125 bits (314), Expect = 8e-29, Method: Compositional matrix adjust.
Identities = 61/178 (34%), Positives = 102/178 (57%), Gaps = 6/178 (3%)
Query 1 KVETCKWNGWGYCDTYVSMSSASMIRLHGSRYSLCGVDMGHWHEFMESIPGIEFAFSSPA 60
+ E KWNGWGY D+ ++ + L G RY L G+ + +++++ G+ + +
Sbjct 100 RQEVMKWNGWGYNDSKFLLNKKGQVELTGKRYPLSGLVLPTLRDWIQNTLGVSLEHKTTS 159
Query 61 QDTLVIP---PQRVNNEAFIKELQQLAPEIEVANSEKERLFHGHGHTCNEIFELRYGTVA 117
+ T + P P + NE F++EL++ I + +R+F HGH +EIF LR G
Sbjct 160 K-TSINPSEAPPSIVNEDFLQELKE--ARISYSQEADDRVFRAHGHCLHEIFLLREGMFE 216
Query 118 RLPDVVLYPKEHKDVEAIVECASRHNVCLIPFGGGTSVTLGVAVPVSESRMVATVSLA 175
R+PD+V++P H DV IV A ++N+C+IP GGGTSV+ G+ P E+R + ++ +
Sbjct 217 RIPDIVVWPTCHDDVVKIVNLACKYNLCIIPIGGGTSVSYGLMCPADETRTIISLDTS 274
> ath:AT5G06580 FAD linked oxidase family protein (EC:1.1.2.4);
K00102 D-lactate dehydrogenase (cytochrome) [EC:1.1.2.4]
Length=567
Score = 42.4 bits (98), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 21/80 (26%), Positives = 40/80 (50%), Gaps = 8/80 (10%)
Query 76 FIKELQQLAPEIEVANSEKERLFHGHGHTCNEIFELRYGTVARLPDVVLYPKEHKDVEAI 135
I +L+ + E + ER FHG + +PDVV++P+ ++V I
Sbjct 110 LISQLKTIL-EDNLTTDYDERYFHGKPQNS-------FHKAVNIPDVVVFPRSEEEVSKI 161
Query 136 VECASRHNVCLIPFGGGTSV 155
++ + + V ++P+GG TS+
Sbjct 162 LKSCNEYKVPIVPYGGATSI 181
> dre:100333035 lactate dehydrogenase D-like
Length=484
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 41/80 (51%), Gaps = 11/80 (13%)
Query 83 LAPEIEVANSEKERLFHGHGHTCNEIFELRYGTVARLPDVVLYPKEHKDVEAIVECASRH 142
LA +++ + +E+ H HT + +LPD V++ + DV+A+V S H
Sbjct 40 LADKLQTGQAFREQ----HTHTTTYL-------TRQLPDAVVFVENSDDVKAVVRACSEH 88
Query 143 NVCLIPFGGGTSVTLGVAVP 162
V +IPFG G+S+ V P
Sbjct 89 GVPIIPFGTGSSLEGQVNAP 108
> tpv:TP01_0575 hypothetical protein
Length=1723
Score = 38.1 bits (87), Expect = 0.017, Method: Composition-based stats.
Identities = 18/44 (40%), Positives = 27/44 (61%), Gaps = 2/44 (4%)
Query 69 QRVNNEAFIKELQQLAPEIEVANSEKERLFHGHGHTCNEIFELR 112
+R+NN KE+ L ++E N+E ERL GH HT E+ +L+
Sbjct 873 ERINNRQ--KEVDALKSDLEAKNAEAERLLSGHLHTKGELEKLK 914
> eco:b1687 ydiJ, ECK1684, JW1677; predicted FAD-linked oxidoreductase;
K06911
Length=1018
Score = 37.7 bits (86), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 26/84 (30%), Positives = 40/84 (47%), Gaps = 13/84 (15%)
Query 76 FIKELQQLAPEIEVANSEKERLFHGHGHTCNEIFELRYGTVARLPDVVLYPKEHKDVEAI 135
F++EL+Q + A S +RL T N I++L LPD V++P+ DV I
Sbjct 18 FLQELEQQGFTGDTATSYADRLTMS---TDNSIYQL-------LPDAVVFPRSTADVALI 67
Query 136 VECASRH---NVCLIPFGGGTSVT 156
A++ ++ P GGGT
Sbjct 68 ARLAAQERYSSLIFTPRGGGTGTN 91
> dre:334208 ldhd, wu:fi36b04, zgc:55447; lactate dehydrogenase
D (EC:1.1.2.4); K00102 D-lactate dehydrogenase (cytochrome)
[EC:1.1.2.4]
Length=497
Score = 37.0 bits (84), Expect = 0.037, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 28/43 (65%), Gaps = 0/43 (0%)
Query 118 RLPDVVLYPKEHKDVEAIVECASRHNVCLIPFGGGTSVTLGVA 160
R PDVV++P+ ++V A+ + + + +IPFG GT + GV+
Sbjct 68 RPPDVVVFPRSVEEVSALAKICHHYRLPIIPFGTGTGLEGGVS 110
> cel:F54D5.12 hypothetical protein
Length=487
Score = 36.6 bits (83), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 33/49 (67%), Gaps = 1/49 (2%)
Query 122 VVLYPKEHKDVEAIVECASRHNVCLIPFGGGTSVTLGVAVPVSESRMVA 170
VVLYPK ++V AI+ S++ + ++P GG T + +G ++PV + +++
Sbjct 69 VVLYPKSTEEVSAILAYCSKNKLAVVPQGGNTGL-VGGSIPVHDEVVIS 116
> sce:YDL174C DLD1; D-lactate dehydrogenase, oxidizes D-lactate
to pyruvate, transcription is heme-dependent, repressed by
glucose, and derepressed in ethanol or lactate; located in
the mitochondrial inner membrane (EC:1.1.2.4); K00102 D-lactate
dehydrogenase (cytochrome) [EC:1.1.2.4]
Length=587
Score = 34.3 bits (77), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 18/82 (21%), Positives = 41/82 (50%), Gaps = 3/82 (3%)
Query 74 EAFIKELQQLAPEIEVANSEKERLFHGHGHTCNEIFELRYGTVARLPDVVLYPKEHKDVE 133
+ +++L+Q+ S+ + H T F + + + P ++L+P ++V
Sbjct 108 DKVVEDLKQVLGNKPENYSDAKSDLDAHSDT---YFNTHHPSPEQRPRIILFPHTTEEVS 164
Query 134 AIVECASRHNVCLIPFGGGTSV 155
I++ +N+ ++PF GGTS+
Sbjct 165 KILKICHDNNMPVVPFSGGTSL 186
> hsa:197257 LDHD, DLD, MGC57726; lactate dehydrogenase D (EC:1.1.2.4);
K00102 D-lactate dehydrogenase (cytochrome) [EC:1.1.2.4]
Length=507
Score = 33.1 bits (74), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 120 PDVVLYPKEHKDVEAIVECASRHNVCLIPFGGGTSVTLGV 159
PD V++P+ + V + R V +IPFG GT + GV
Sbjct 66 PDAVVWPQNVEQVSRLAALCYRQGVPIIPFGTGTGLEGGV 105
> sce:YJL141C YAK1; Serine-threonine protein kinase that is part
of a glucose-sensing system involved in growth control in
response to glucose availability; translocates from the cytoplasm
to the nucleus and phosphorylates Pop2p in response to
a glucose signal (EC:2.7.12.1); K00924 [EC:2.7.1.-]
Length=807
Score = 33.1 bits (74), Expect = 0.55, Method: Composition-based stats.
Identities = 31/104 (29%), Positives = 52/104 (50%), Gaps = 17/104 (16%)
Query 62 DTLVIPPQRV-----NNEAFIKELQQLAPEIEVANSEKERLFHGHGHTCNE--IFEL--- 111
DTL PP + N+ F+K +LAPE ++++K R+ C E I E
Sbjct 584 DTLGYPPSWMIDMGKNSGKFMK---KLAPEESSSSTQKHRM-KTIEEFCREYNIVEKPSK 639
Query 112 RYGTVARLPDVVL---YPKEHKDVEAIVECASRHNVCLIPFGGG 152
+Y +LPD++ YPK ++ + +++ ++ CLI F GG
Sbjct 640 QYFKWRKLPDIIRNYRYPKSIQNSQELIDQEMQNRECLIHFLGG 683
> tpv:TP04_0229 hypothetical protein; K14843 pescadillo
Length=457
Score = 32.7 bits (73), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 25/91 (27%), Positives = 40/91 (43%), Gaps = 5/91 (5%)
Query 58 SPAQDTLVIPPQRVNNEAFIKELQQLAPEIEVANSEKERLFHGHGHTCN-EIFELRYGTV 116
SP +D +P ++V E I+E PE E + +K+ N E+ +LR +
Sbjct 367 SPFEDGAYVPDRKVELEKIIREATGKKPEEEGGDKKKDSYIEDLSEELNPELRKLRRSLM 426
Query 117 ----ARLPDVVLYPKEHKDVEAIVECASRHN 143
RL + Y KE K+ EA + + N
Sbjct 427 PKKHKRLLSKIEYAKERKEAEARKLASRKKN 457
> hsa:154664 ABCA13, DKFZp313D2411, FLJ16398, FLJ33876, FLJ33951;
ATP-binding cassette, sub-family A (ABC1), member 13; K05647
ATP binding cassette, subfamily A (ABC1), member 13
Length=5058
Score = 32.3 bits (72), Expect = 0.87, Method: Composition-based stats.
Identities = 13/20 (65%), Positives = 17/20 (85%), Gaps = 0/20 (0%)
Query 68 PQRVNNEAFIKELQQLAPEI 87
P++VNN AF+KE+Q LA EI
Sbjct 106 PKKVNNLAFLKEIQDLAEEI 125
> dre:565889 d2hgdh, zgc:158661; D-2-hydroxyglutarate dehydrogenase
(EC:1.1.99.-)
Length=533
Score = 32.0 bits (71), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 32/57 (56%), Gaps = 1/57 (1%)
Query 115 TVARLPDVVLYPKEHKDVEAIVECASRHNVCLIPFGGGTSVTLGVAVPVSESRMVAT 171
TV DV+L PK + V I+ + N+ + P GG T + +G +VPV + +++T
Sbjct 106 TVQGSSDVLLRPKTTEGVSQILRYCNERNLAVCPQGGNTGL-VGGSVPVFDEIILST 161
> cel:K06G5.2 cyp-13B2; CYtochrome P450 family member (cyp-13B2)
Length=511
Score = 31.2 bits (69), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 13/35 (37%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 133 EAIVECASRHNVCLIPFGGGTSVTLGVAVPVSESR 167
E +E + RHN+ IPFG G +G+ + +SE++
Sbjct 434 ERWLESSPRHNMSWIPFGAGPRQCVGMRLGLSEAK 468
> hsa:23499 MACF1, ABP620, ACF7, FLJ45612, FLJ46776, KIAA0465,
KIAA1251, MACF, OFC4; microtubule-actin crosslinking factor
1
Length=5430
Score = 31.2 bits (69), Expect = 2.1, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 27/48 (56%), Gaps = 1/48 (2%)
Query 41 HWHEFMESIPGIEFAFSSPAQDTLVIPPQRVNNEAFIKELQQLAPEIE 88
W E E + A SSP + T + Q +N+AF+ EL+Q +P+I+
Sbjct 2412 QWLESKEEVLKSMDAMSSPTK-TETVKAQAESNKAFLAELEQNSPKIQ 2458
> dre:793570 btr07p; bloodthirsty-related gene family, member
7, pseudogene
Length=552
Score = 31.2 bits (69), Expect = 2.1, Method: Composition-based stats.
Identities = 18/69 (26%), Positives = 36/69 (52%), Gaps = 3/69 (4%)
Query 73 NEAFIKELQQLAPEIEVANSEKERLFHGHGHTCNEIFELRYGTVARLPDVVLYPKEHKDV 132
+E I+ELQQ E+++ NS+ + L H H ++ E+ Y ++ P++ +P+
Sbjct 280 DEELIQELQQEINELKMRNSDLDHLSHTEDHL--QLLEI-YPSICSPPNIKNWPETSMST 336
Query 133 EAIVECASR 141
+ +E R
Sbjct 337 DVSLETLRR 345
> ath:AT4G36400 FAD linked oxidase family protein
Length=559
Score = 30.8 bits (68), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 40/77 (51%), Gaps = 9/77 (11%)
Query 93 EKERLFHGHGHTCNEIFELRYGTVARLPDVVLYPKEHKDVEAIVECASRHNVCLIPFGGG 152
+KERL T N + +Y ++L +L PK ++V I+E + ++P GG
Sbjct 115 DKERL-----ETANTDWMHKYKGSSKL---MLLPKNTQEVSQILEYCDSRRLAVVPQGGN 166
Query 153 TSVTLGVAVPVSESRMV 169
T + +G +VPV + +V
Sbjct 167 TGL-VGGSVPVFDEVIV 182
> mmu:20495 Slc12a1, AI788571, D630042G03Rik, Nkcc2, mBSC1, urehr3;
solute carrier family 12, member 1; K14425 solute carrier
family 12 (sodium/potassium/chloride transporter), member
1
Length=1095
Score = 30.4 bits (67), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 17/65 (26%), Positives = 34/65 (52%), Gaps = 5/65 (7%)
Query 43 HEFMESIPGIEFAFSSPAQDTLVIPPQRVNNEAFIKELQQLAPEIEVANSEKERLFHGHG 102
H M+++P IE+ ++T + +VN + ++ +QLA + VA +R+ +G G
Sbjct 99 HNTMDAVPKIEYY-----RNTGSVSGPKVNRPSLLEIHEQLAKNVTVAPGSADRVANGDG 153
Query 103 HTCNE 107
+E
Sbjct 154 MPGDE 158
> hsa:6557 SLC12A1, BSC1, MGC48843, NKCC2; solute carrier family
12 (sodium/potassium/chloride transporters), member 1; K14425
solute carrier family 12 (sodium/potassium/chloride transporter),
member 1
Length=1099
Score = 30.4 bits (67), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 17/60 (28%), Positives = 32/60 (53%), Gaps = 5/60 (8%)
Query 43 HEFMESIPGIEFAFSSPAQDTLVIPPQRVNNEAFIKELQQLAPEIEVANSEKERLFHGHG 102
H M+++P IE+ ++T I +VN + ++ +QLA + V S +R+ +G G
Sbjct 103 HNTMDAVPKIEYY-----RNTGSISGPKVNRPSLLEIHEQLAKNVAVTPSSADRVANGDG 157
> sce:YLL035W GRC3; Grc3p; K06947
Length=632
Score = 30.4 bits (67), Expect = 3.5, Method: Composition-based stats.
Identities = 21/69 (30%), Positives = 31/69 (44%), Gaps = 6/69 (8%)
Query 42 WHEFMESIPGIEFAFSSPAQDTLVIPPQRVNNEAFIK--ELQQLAPEIEVANSEKERLFH 99
WH +SIP I+F+ + QDT +P NN I+ E + + V NS L
Sbjct 121 WHPLSQSIPTIDFSHFAGWQDTFFMPR---NNRFKIRDEEFKSFPCVLRVFNSNHTGLLE 177
Query 100 GHGHTCNEI 108
GH ++
Sbjct 178 A-GHLYRDV 185
> cel:F55F10.3 hypothetical protein
Length=181
Score = 30.0 bits (66), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 13/43 (30%), Positives = 24/43 (55%), Gaps = 2/43 (4%)
Query 35 CGVDMGHWHEFMESIPGIEFAFSSPAQDTLVIPPQRVNNEAFI 77
C +D+ HW + ++ +EF +SPA+ V+PP + E +
Sbjct 50 CLIDIKHWRKSIDGRAQLEFPSASPAE--FVLPPIQRTPENLV 90
> hsa:7011 TEP1, TLP1, TP1, TROVE1, VAULT2, p240; telomerase-associated
protein 1; K11127 telomerase protein component 1
Length=2627
Score = 30.0 bits (66), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 23/49 (46%), Gaps = 0/49 (0%)
Query 98 FHGHGHTCNEIFELRYGTVARLPDVVLYPKEHKDVEAIVECASRHNVCL 146
H H T ++I L+ +A LPD+ K H V A + S N CL
Sbjct 34 LHQHVSTHSDILSLKNQCLATLPDLKTMEKPHGYVSAHPDILSLENQCL 82
> cel:F02C12.5 cyp-13B1; CYtochrome P450 family member (cyp-13B1);
K00517 [EC:1.14.-.-]
Length=510
Score = 30.0 bits (66), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 13/35 (37%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 133 EAIVECASRHNVCLIPFGGGTSVTLGVAVPVSESR 167
E +E +SRH + IPFG G +G+ + +SE++
Sbjct 433 ERWLEPSSRHTMSWIPFGAGPRQCVGMRLGLSEAK 467
> hsa:286365 OR13D1, OR9-15; olfactory receptor, family 13, subfamily
D, member 1; K04257 olfactory receptor
Length=346
Score = 29.3 bits (64), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 27/43 (62%), Gaps = 1/43 (2%)
Query 101 HGHTCNEIFELRYGTVARLPDVVLYPKEHKDV-EAIVECASRH 142
+ +T +EI L YG V+ + + ++Y +K+V EA+ + SRH
Sbjct 298 NTNTSDEIIGLSYGVVSPMLNPIIYSLRNKEVKEAVKKVLSRH 340
> cpv:cgd5_2440 Ylr401cp-like protein with 2 CCCH domains plus
Dus1p tRNA dihydrouridine synthase Tim barrel ; K05544 tRNA-dihydrouridine
synthase 3 [EC:1.-.-.-]
Length=707
Score = 28.9 bits (63), Expect = 8.8, Method: Composition-based stats.
Identities = 20/76 (26%), Positives = 33/76 (43%), Gaps = 5/76 (6%)
Query 70 RVNNEAFIKELQQLAPEIEVANSEKERLFHGHGHTCNEIFELRYGTVARLPDVVLYPKEH 129
+ NE I E+++L + ANS K+R G L Y +A++ D+ +
Sbjct 41 KCQNETGISEVKKLKEDQNEANSSKKRSIRGSFKASERSQNLNY--IAKMDDL---DRLC 95
Query 130 KDVEAIVECASRHNVC 145
++ EC HN C
Sbjct 96 SNIAIFGECKLDHNTC 111
Lambda K H
0.320 0.135 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4600750868
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40