bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1173_orf1
Length=78
Score E
Sequences producing significant alignments: (Bits) Value
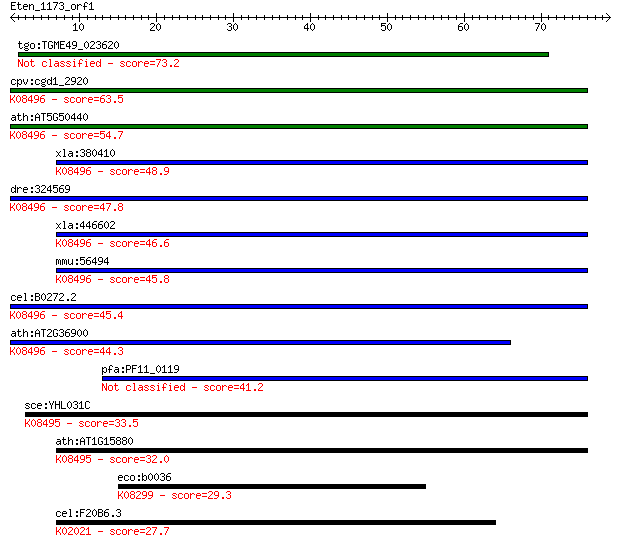
tgo:TGME49_023620 golgi SNARE protein, putative 73.2 2e-13
cpv:cgd1_2920 golgi transport SNARE BOS1 secretory pathway pro... 63.5 1e-10
ath:AT5G50440 MEMB12; MEMB12 (MEMBRIN 12); SNAP receptor; K084... 54.7 6e-08
xla:380410 gosr2, MGC64459; golgi SNAP receptor complex member... 48.9 4e-06
dre:324569 gosr2, wu:fc33b02, zgc:64068; golgi SNAP receptor c... 47.8 9e-06
xla:446602 MGC81773 protein; K08496 golgi SNAP receptor comple... 46.6 2e-05
mmu:56494 Gosr2, 2310032N09Rik, C76855, Gs27, SNARE, membrin; ... 45.8 3e-05
cel:B0272.2 gosr-2.1; GOlgi Snap Receptor complex member famil... 45.4 5e-05
ath:AT2G36900 MEMB11; MEMB11 (MEMBRIN 11); SNAP receptor; K084... 44.3 1e-04
pfa:PF11_0119 GS27; SNARE protein, putative 41.2 8e-04
sce:YHL031C GOS1; Gos1p; K08495 golgi SNAP receptor complex me... 33.5 0.19
ath:AT1G15880 GOS11; GOS11 (golgi snare 11); SNARE binding; K0... 32.0 0.48
eco:b0036 caiD, ECK0037, JW0035, yaaL; carnitinyl-CoA dehydrat... 29.3 2.9
cel:F20B6.3 mrp-6; Multidrug Resistance Protein family member ... 27.7 8.8
> tgo:TGME49_023620 golgi SNARE protein, putative
Length=264
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 34/69 (49%), Positives = 56/69 (81%), Gaps = 0/69 (0%)
Query 2 DSRTVLDSVLSQGWGIVGQLVSQNASLKNARRKVLDMASSAGVASTLLGAVGRRQQGDKW 61
+S ++LD++LSQG + ++V QN+ LK+A+RK+LDM +S+G+A++LLGA+ RR+ D+
Sbjct 165 ESHSMLDAILSQGRSALDKVVQQNSVLKSAKRKILDMNTSSGLAASLLGAISRREATDRR 224
Query 62 LVYGGMVVT 70
LV+GGM +T
Sbjct 225 LVWGGMFLT 233
> cpv:cgd1_2920 golgi transport SNARE BOS1 secretory pathway protein,
possible transmembrane or GPI anchor at C-terminus ;
K08496 golgi SNAP receptor complex member 2
Length=220
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 31/75 (41%), Positives = 45/75 (60%), Gaps = 0/75 (0%)
Query 1 QDSRTVLDSVLSQGWGIVGQLVSQNASLKNARRKVLDMASSAGVASTLLGAVGRRQQGDK 60
Q+S +D L Q IV L +QN LK+ R+K LDMAS G++ +LL + RR D+
Sbjct 134 QESHIAIDHALEQARSIVFNLKNQNRMLKSVRKKALDMASRLGISHSLLSNIERRNLVDQ 193
Query 61 WLVYGGMVVTLIFFF 75
LVYG +++T + F
Sbjct 194 ILVYGCILLTSLIFI 208
> ath:AT5G50440 MEMB12; MEMB12 (MEMBRIN 12); SNAP receptor; K08496
golgi SNAP receptor complex member 2
Length=219
Score = 54.7 bits (130), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 46/75 (61%), Gaps = 0/75 (0%)
Query 1 QDSRTVLDSVLSQGWGIVGQLVSQNASLKNARRKVLDMASSAGVASTLLGAVGRRQQGDK 60
++S+ +L+ G I+ + Q LK+A+RK LD+ ++ G+++++L + RR + D
Sbjct 138 KNSKRMLEDSFQSGVAILSKYAEQRDRLKSAQRKALDVLNTVGLSNSVLRLIERRNRVDT 197
Query 61 WLVYGGMVVTLIFFF 75
W+ Y GM+ TL+ +
Sbjct 198 WIKYAGMIATLVILY 212
> xla:380410 gosr2, MGC64459; golgi SNAP receptor complex member
2; K08496 golgi SNAP receptor complex member 2
Length=210
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 43/69 (62%), Gaps = 0/69 (0%)
Query 7 LDSVLSQGWGIVGQLVSQNASLKNARRKVLDMASSAGVASTLLGAVGRRQQGDKWLVYGG 66
+D +L G I+ L Q +LK A +K+LD+A+ G+++T++ + +R DK ++ GG
Sbjct 135 IDDLLGSGSSILEGLRDQRKTLKGAHKKILDVANMLGLSNTVMRLIEKRAFQDKVIMIGG 194
Query 67 MVVTLIFFF 75
M++T +F
Sbjct 195 MLLTCVFMI 203
> dre:324569 gosr2, wu:fc33b02, zgc:64068; golgi SNAP receptor
complex member 2; K08496 golgi SNAP receptor complex member
2
Length=212
Score = 47.8 bits (112), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 45/75 (60%), Gaps = 0/75 (0%)
Query 1 QDSRTVLDSVLSQGWGIVGQLVSQNASLKNARRKVLDMASSAGVASTLLGAVGRRQQGDK 60
Q++ +D +L G I+ L Q ++LK +K+LD+A+ G+++T++ + +R DK
Sbjct 131 QNAHRGMDDLLGSGSSILNGLRDQRSTLKGTHKKMLDVANMLGLSNTVMRLIEKRASQDK 190
Query 61 WLVYGGMVVTLIFFF 75
+++ GM+ T + F
Sbjct 191 FIMMAGMLATCVVMF 205
> xla:446602 MGC81773 protein; K08496 golgi SNAP receptor complex
member 2
Length=194
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 42/69 (60%), Gaps = 0/69 (0%)
Query 7 LDSVLSQGWGIVGQLVSQNASLKNARRKVLDMASSAGVASTLLGAVGRRQQGDKWLVYGG 66
+D +L G ++ L Q +LK +K+LD+A+ G+++T++ + +R DK ++ GG
Sbjct 119 IDDLLGSGSSVLEGLRDQRKTLKGTHKKILDVANMLGLSNTVMRLIEKRAFQDKAIMIGG 178
Query 67 MVVTLIFFF 75
M++T +F
Sbjct 179 MLLTCVFMI 187
> mmu:56494 Gosr2, 2310032N09Rik, C76855, Gs27, SNARE, membrin;
golgi SNAP receptor complex member 2; K08496 golgi SNAP receptor
complex member 2
Length=212
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 43/69 (62%), Gaps = 0/69 (0%)
Query 7 LDSVLSQGWGIVGQLVSQNASLKNARRKVLDMASSAGVASTLLGAVGRRQQGDKWLVYGG 66
+D ++ G I+ L +Q +LK ++K+LD+A+ G+++T++ + +R DK+ + GG
Sbjct 137 MDDLIGGGHSILEGLRAQRLTLKGTQKKILDIANMLGLSNTVMRLIEKRAFQDKYFMIGG 196
Query 67 MVVTLIFFF 75
M++T F
Sbjct 197 MLLTCAVMF 205
> cel:B0272.2 gosr-2.1; GOlgi Snap Receptor complex member family
member (gosr-2.1); K08496 golgi SNAP receptor complex member
2
Length=213
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 24/75 (32%), Positives = 43/75 (57%), Gaps = 0/75 (0%)
Query 1 QDSRTVLDSVLSQGWGIVGQLVSQNASLKNARRKVLDMASSAGVASTLLGAVGRRQQGDK 60
Q S T LD ++SQG ++ L SQ+ +L+ RK+ ++ + G++++ L + RR + D
Sbjct 131 QSSHTHLDDLISQGSAVLENLKSQHLNLRGVGRKMHEIGQALGLSNSTLQVIDRRVREDW 190
Query 61 WLVYGGMVVTLIFFF 75
L G +V IF +
Sbjct 191 ILFVIGCIVCCIFMY 205
> ath:AT2G36900 MEMB11; MEMB11 (MEMBRIN 11); SNAP receptor; K08496
golgi SNAP receptor complex member 2
Length=225
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 40/65 (61%), Gaps = 0/65 (0%)
Query 1 QDSRTVLDSVLSQGWGIVGQLVSQNASLKNARRKVLDMASSAGVASTLLGAVGRRQQGDK 60
++S+ +L+ S G I+ + Q LK+A+RK LD+ ++ G+++++L + RR + D
Sbjct 144 KNSKRMLEESFSSGVAILSKYAEQRDRLKSAQRKALDVLNTVGLSNSVLRLIERRNRVDT 203
Query 61 WLVYG 65
W+ Y
Sbjct 204 WIKYA 208
> pfa:PF11_0119 GS27; SNARE protein, putative
Length=289
Score = 41.2 bits (95), Expect = 8e-04, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 40/63 (63%), Gaps = 0/63 (0%)
Query 13 QGWGIVGQLVSQNASLKNARRKVLDMASSAGVASTLLGAVGRRQQGDKWLVYGGMVVTLI 72
QG + L QN LKN R+KV+DM + G++S+L+ + + + + +V G++++LI
Sbjct 219 QGMNTLNMLRKQNKFLKNVRKKVIDMYNYVGLSSSLVDTIKKAHKQNLIIVIVGIILSLI 278
Query 73 FFF 75
FF+
Sbjct 279 FFY 281
> sce:YHL031C GOS1; Gos1p; K08495 golgi SNAP receptor complex
member 1
Length=223
Score = 33.5 bits (75), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 18/73 (24%), Positives = 34/73 (46%), Gaps = 0/73 (0%)
Query 3 SRTVLDSVLSQGWGIVGQLVSQNASLKNARRKVLDMASSAGVASTLLGAVGRRQQGDKWL 62
S V+D ++SQ W Q SQ+ L A KVL + L+ + R++ + ++
Sbjct 148 SNNVVDRLISQAWETRSQFHSQSNVLNTANNKVLQTLQRIPGVNQLIMKINTRRKKNAFV 207
Query 63 VYGGMVVTLIFFF 75
+ + ++F F
Sbjct 208 LATITTLCILFLF 220
> ath:AT1G15880 GOS11; GOS11 (golgi snare 11); SNARE binding;
K08495 golgi SNAP receptor complex member 1
Length=223
Score = 32.0 bits (71), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 19/71 (26%), Positives = 36/71 (50%), Gaps = 2/71 (2%)
Query 7 LDSVLSQGWGIVGQLVSQNASLKNARRKVLDMASSAGVASTLLGAVGRRQQGDKWL--VY 64
+D V+SQ +G LV Q ++ K+ ++AS +T+L A+ R++ D + +
Sbjct 149 MDGVISQAQATLGTLVFQRSTFGGINSKLSNVASRLPTVNTILAAIKRKKSMDTIILSLV 208
Query 65 GGMVVTLIFFF 75
+ LIF +
Sbjct 209 AAVCTFLIFIY 219
> eco:b0036 caiD, ECK0037, JW0035, yaaL; carnitinyl-CoA dehydratase
(EC:4.2.1.-); K08299 carnitinyl-CoA dehydratase [EC:4.2.1.-]
Length=261
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 15 WGIVGQLVSQNASLKNARRKVLDMASSAGVASTLLGAVGR 54
WGIV ++VSQ + NAR + +SA +A L + R
Sbjct 173 WGIVNRVVSQAELMDNARELAQQLVNSAPLAIAALKEIYR 212
> cel:F20B6.3 mrp-6; Multidrug Resistance Protein family member
(mrp-6); K02021 putative ABC transport system ATP-binding
protein
Length=1396
Score = 27.7 bits (60), Expect = 8.8, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 24/57 (42%), Gaps = 0/57 (0%)
Query 7 LDSVLSQGWGIVGQLVSQNASLKNARRKVLDMASSAGVASTLLGAVGRRQQGDKWLV 63
LD L W G++ +N S+ KVL S + +G VGR G L+
Sbjct 1122 LDKQLLNSWPTKGEIEFENVSVNYGETKVLKNLSFVVLGGQKIGVVGRTGAGKSTLI 1178
Lambda K H
0.321 0.134 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2067704464
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40