bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
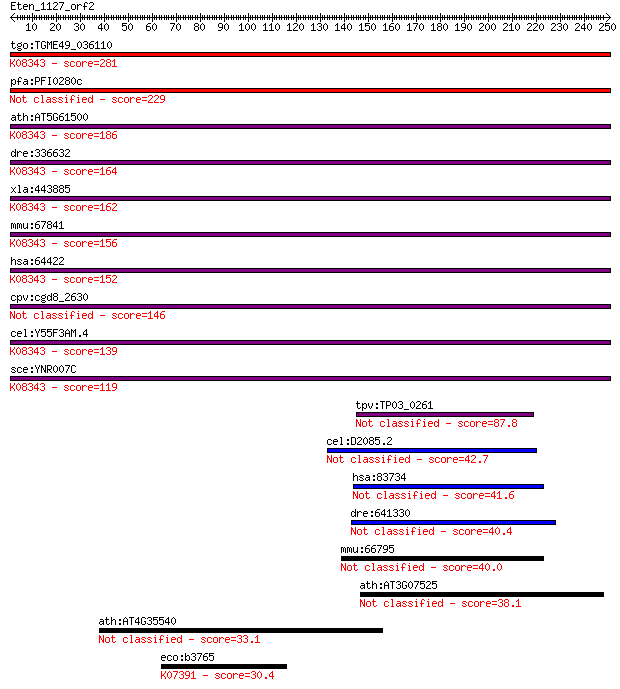
Query= Eten_1127_orf2
Length=250
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_036110 autophagocytosis associated protein, putativ... 281 1e-75
pfa:PFI0280c autophagocytosis associated protein, putative 229 5e-60
ath:AT5G61500 ATG3; ATG3; K08343 autophagy-related protein 3 186 6e-47
dre:336632 apg3l, atg3, wu:fb15g12, zgc:64156; APG3 autophagy ... 164 2e-40
xla:443885 atg3, apg3l; ATG3 autophagy related 3 homolog; K083... 162 1e-39
mmu:67841 Atg3, 2610016C12Rik, APG3, Apg3l, Atg3l, PC3-96; aut... 156 7e-38
hsa:64422 ATG3, APG3, APG3-LIKE, APG3L, DKFZp564M1178, FLJ2212... 152 1e-36
cpv:cgd8_2630 APG10/ Aut1p like like autophagocytosis protein ... 146 7e-35
cel:Y55F3AM.4 atg-3; AuTophaGy (yeast Atg homolog) family memb... 139 1e-32
sce:YNR007C ATG3, APG3, AUT1; Atg3p; K08343 autophagy-related ... 119 8e-27
tpv:TP03_0261 hypothetical protein 87.8 3e-17
cel:D2085.2 atg-10; AuTophaGy (yeast Atg homolog) family membe... 42.7 0.001
hsa:83734 ATG10, APG10, APG10L, DKFZp586I0418, FLJ13954, pp126... 41.6 0.003
dre:641330 atg10, MGC123341, im:7153729, zgc:123341; ATG10 aut... 40.4 0.007
mmu:66795 Atg10, 5330424L23Rik, 5430428K15Rik, AI852123, APG10... 40.0 0.009
ath:AT3G07525 ATG10; ATG10 (autophagy 10); transporter/ ubiqui... 38.1 0.034
ath:AT4G35540 transcription regulator/ zinc ion binding 33.1 1.1
eco:b3765 yifB, ECK3758, JW3738; predicted bifunctional enzyme... 30.4 5.7
> tgo:TGME49_036110 autophagocytosis associated protein, putative
; K08343 autophagy-related protein 3
Length=397
Score = 281 bits (720), Expect = 1e-75, Method: Compositional matrix adjust.
Identities = 148/275 (53%), Positives = 187/275 (68%), Gaps = 27/275 (9%)
Query 1 DLLVFKFPTWQWEPA--VGKRINSWLPPDKQFLVTRGVACHRRVRDIEASLAAEAREDAE 58
DLL KFPTWQW+ GKR + WLP DKQ+L+T+ V C+RRVRD++ +L D E
Sbjct 105 DLLTHKFPTWQWKGVGPTGKRASGWLPEDKQYLITKNVPCYRRVRDMDDALNTRVGHDVE 164
Query 59 G-WVLPVEAEEPEEV--------LSVSVSGGETNP-----LEKPP--------GGSGSSL 96
G W+LP+ +E E L+ S+ N L KP + S+
Sbjct 165 GGWMLPLLNDEEREGGSSGEAPDLTQSMQNLRLNAECGHELRKPAPPTPTQTSASTASNQ 224
Query 97 RDTSGGALPDLKNLRDLDSLLCEDD-PACADSSTSYFVRNAPDADVLSVRTYDLSITYDK 155
++ +PDL N D+D L+ EDD PA A++ + VR +PDA++++ R+YDLSITYDK
Sbjct 225 ENSLRDDVPDLINFADIDCLVQEDDDPAAAEAPS--VVRTSPDAEIVAARSYDLSITYDK 282
Query 156 YFQTPRLWLFGYNESGTPLTPVEIFEDILTDYAAKTVTVDPHPCTGVPTASIHPCKHAQV 215
YFQTPR+WLFGY+E+G PL P EIFEDILTDYAAKTVTVDPHPCTG+PTASIHPC+HA V
Sbjct 283 YFQTPRIWLFGYSENGVPLLPEEIFEDILTDYAAKTVTVDPHPCTGIPTASIHPCRHASV 342
Query 216 MKKVVDHWRSQGVEPRPDLALIVLLKFISSVVPTI 250
MKKVVD W GV PR DLAL++LLKF+SSV+PTI
Sbjct 343 MKKVVDSWVESGVRPRHDLALLILLKFVSSVIPTI 377
> pfa:PFI0280c autophagocytosis associated protein, putative
Length=313
Score = 229 bits (585), Expect = 5e-60, Method: Compositional matrix adjust.
Identities = 116/252 (46%), Positives = 163/252 (64%), Gaps = 5/252 (1%)
Query 1 DLLVFKFPTWQWEPAVGKRINSWLPPDKQFLVTRGVACHRRVRDIEASLAAEAREDAEGW 60
D LV+KF TW+W+ A R+ +LP +KQFL+T+ V C +R++D+ ++ + + W
Sbjct 47 DFLVYKFKTWEWQEADKDRVVPYLPENKQFLITKNVPCKQRIKDLN-NIVHDLKIVDNDW 105
Query 61 VLPVEAEE--PEEVLSVSVSGGETNPLEKPPGGSGSSLRDTSGGALPDLKNLRDLDSLLC 118
+LP E+ P ++ + E +K D + D+ N ++L+
Sbjct 106 LLPSYEEDNNPTDIYEY-LPNSEYTINDKNIYNYEEEEEDDNCDEAIDINNFYMENNLIK 164
Query 119 EDDPACADSSTSYFVRNAPDADVLSVRTYDLSITYDKYFQTPRLWLFGYNESGTPLTPVE 178
E DPA +S TS + +N +++ +RTYD+SITYDKY+QTPR+WLFGYNE+G PL E
Sbjct 165 EHDPASINS-TSCYSKNMLHDNLMKIRTYDVSITYDKYYQTPRIWLFGYNENGDPLKSEE 223
Query 179 IFEDILTDYAAKTVTVDPHPCTGVPTASIHPCKHAQVMKKVVDHWRSQGVEPRPDLALIV 238
IFEDIL+DY+ KTVT DPHPCTGV TASIHPCKHA+ + VV++W S+ EPR DL L+
Sbjct 224 IFEDILSDYSYKTVTYDPHPCTGVMTASIHPCKHAEAILNVVNNWISEEKEPRHDLYLLF 283
Query 239 LLKFISSVVPTI 250
LLKFIS V+PTI
Sbjct 284 LLKFISGVIPTI 295
> ath:AT5G61500 ATG3; ATG3; K08343 autophagy-related protein 3
Length=313
Score = 186 bits (472), Expect = 6e-47, Method: Compositional matrix adjust.
Identities = 109/263 (41%), Positives = 150/263 (57%), Gaps = 20/263 (7%)
Query 1 DLLVFKFPTWQWEPAVGKRINSWLPPDKQFLVTRGVACHRRVRDIEASLAAEA------R 54
D LV K PTW WE + +LP DKQFL+TR V C RR + A
Sbjct 43 DNLVSKCPTWSWESGDASKRKPYLPSDKQFLITRNVPCLRRAASVAEDYEAAGGEVLVDD 102
Query 55 EDAEGWVL----PVEAEEPEEVLSV--SVSGGETNPLEKPPGGSGSSLRDTSGGALPDLK 108
ED +GW+ P + + E+ L ++ E N ++ P G D +PD++
Sbjct 103 EDNDGWLATHGKPKDKGKEEDNLPSMDALDINEKNTIQSIPTYFGGEEDDD----IPDME 158
Query 109 NLRDLDSLLCEDDPACADSSTSYFVRNAPDAD-VLSVRTYDLSITYDKYFQTPRLWLFGY 167
+ D+++ E+DPA S+ Y V + PD D +L RTYDLSITYDKY+QTPR+WL GY
Sbjct 159 EFDEADNVV-ENDPATLQST--YLVAHEPDDDNILRTRTYDLSITYDKYYQTPRVWLTGY 215
Query 168 NESGTPLTPVEIFEDILTDYAAKTVTVDPHPCTGVPTASIHPCKHAQVMKKVVDHWRSQG 227
+ES L P + ED+ D+A KTVT++ HP AS+HPC+H VMKK++D S+G
Sbjct 216 DESRMLLQPELVMEDVSQDHARKTVTIEDHPHLPGKHASVHPCRHGAVMKKIIDVLMSRG 275
Query 228 VEPRPDLALIVLLKFISSVVPTI 250
VEP D L + LKF++SV+PTI
Sbjct 276 VEPEVDKYLFLFLKFMASVIPTI 298
> dre:336632 apg3l, atg3, wu:fb15g12, zgc:64156; APG3 autophagy
3-like (EC:6.3.2.-); K08343 autophagy-related protein 3
Length=317
Score = 164 bits (416), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 101/265 (38%), Positives = 140/265 (52%), Gaps = 16/265 (6%)
Query 1 DLLVFKFPTWQWEPAVGKRINSWLPPDKQFLVTRGVACHRRVRDIEASLAAEA---REDA 57
D LV PTW+W ++ +LP DKQFL+TR V C++R + +E S EA +D
Sbjct 44 DHLVHHCPTWKWASGEEAKVKPYLPNDKQFLLTRNVPCYKRCKQMEYSDELEAIIEEDDG 103
Query 58 EG-WVLPVEAEE----PEEVLSVSVSGGETNPLEKPPGGSGSSLRDTSGGALPDLKNLRD 112
+G WV E V +S+ + + G G+S D
Sbjct 104 DGGWVDTFHNSGVTGVTEAVREISLDNKDNMNMNVKTGACGNSGDDDDDEEGEAADMEEY 163
Query 113 LDSLLCEDDPAC------ADSSTSYFVRNAPDADVLSVRTYDLSITYDKYFQTPRLWLFG 166
+S L E D A AD S + DA +L RTYDL ITYDKY+QTPRLWLFG
Sbjct 164 EESGLLETDDATLDTSKMADLSKTKAEAGGEDA-ILQTRTYDLYITYDKYYQTPRLWLFG 222
Query 167 YNESGTPLTPVEIFEDILTDYAAKTVTVDPHPCTGVPT-ASIHPCKHAQVMKKVVDHWRS 225
Y+E PLT +++EDI D+ KTVT++ HP P S+HPC+HA+VMKK+++
Sbjct 223 YDEDRQPLTVDQMYEDISQDHVKKTVTIENHPNLPPPAMCSVHPCRHAEVMKKIIETVAE 282
Query 226 QGVEPRPDLALIVLLKFISSVVPTI 250
G E + L++ LKF+ +V+PTI
Sbjct 283 GGGELGVHMYLLIFLKFVQAVIPTI 307
> xla:443885 atg3, apg3l; ATG3 autophagy related 3 homolog; K08343
autophagy-related protein 3
Length=313
Score = 162 bits (409), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 104/271 (38%), Positives = 147/271 (54%), Gaps = 32/271 (11%)
Query 1 DLLVFKFPTWQWEPAVGKRINSWLPPDKQFLVTRGVACHRRVRDIEASLAAEA---REDA 57
D LV PTWQW +I +LP DKQFL+T+ V C++R + +E S EA +D
Sbjct 44 DHLVHHCPTWQWSAGEESKIKPYLPNDKQFLMTKNVPCYKRCKQMEYSDEQEAIIEEDDG 103
Query 58 EG-WV-------LPVEAEEPEEV-LSVSVSGGETNPLEKPPGGSGSSLRDTSGGALPDLK 108
+G WV L E +E+ L G T+ + + D G D++
Sbjct 104 DGGWVDTFHHTGLSGVTEAVKEITLETQDCGKTTDNI------AVCDDDDDDEGEAADME 157
Query 109 NLRDLDSLLCEDDPACADSSTSYFVRNA--PDAD------VLSVRTYDLSITYDKYFQTP 160
+ + S L E+D A D+S ++ A P AD +L RTYDL ITYDKY+QTP
Sbjct 158 DYEE--SGLLENDDATVDTSK---IKEACKPKADLGGEDAILQTRTYDLYITYDKYYQTP 212
Query 161 RLWLFGYNESGTPLTPVEIFEDILTDYAAKTVTVDPHPCT-GVPTASIHPCKHAQVMKKV 219
RLWLFGY+E PL ++EDI D+ KTVT++ HP P S+HPC+HA+VMKK+
Sbjct 213 RLWLFGYDEQRRPLAVENMYEDISQDHVKKTVTIENHPHLPPPPMCSVHPCRHAEVMKKI 272
Query 220 VDHWRSQGVEPRPDLALIVLLKFISSVVPTI 250
++ G E + L++ LKF+ +V+PTI
Sbjct 273 IETVAEGGGELGVHMYLLIFLKFVQAVIPTI 303
> mmu:67841 Atg3, 2610016C12Rik, APG3, Apg3l, Atg3l, PC3-96; autophagy-related
3 (yeast); K08343 autophagy-related protein
3
Length=314
Score = 156 bits (394), Expect = 7e-38, Method: Compositional matrix adjust.
Identities = 101/270 (37%), Positives = 144/270 (53%), Gaps = 29/270 (10%)
Query 1 DLLVFKFPTWQWEPAVGKRINSWLPPDKQFLVTRGVACHRRVRDIEASLAAEA---REDA 57
D LV PTWQW ++ ++LP DKQFLVT+ V C++R + +E S EA +D
Sbjct 44 DHLVHHCPTWQWATGEELKVKAYLPTDKQFLVTKNVPCYKRCKQMEYSDELEAIIEEDDG 103
Query 58 EG-WVLPVEAEEPEEVLSVSVSGGETNPLEKPPGGSGSSLRDTSGGALPDLKNLRDL--- 113
+G WV + + G T +++ S S++ AL D ++ D
Sbjct 104 DGGWV---------DTYHNTGITGITEAVKEITLESKDSIKLQDCSALCDEEDEEDEGEA 154
Query 114 -------DSLLCEDDPACADS-STSYFVRNAPDAD----VLSVRTYDLSITYDKYFQTPR 161
+S L E D A D+ + DA +L RTYDL ITYDKY+QTPR
Sbjct 155 ADMEEYEESGLLETDEATLDTRKIVEACKAKADAGGEDAILQTRTYDLYITYDKYYQTPR 214
Query 162 LWLFGYNESGTPLTPVEIFEDILTDYAAKTVTVDPHPCT-GVPTASIHPCKHAQVMKKVV 220
LWLFGY+E PLT ++EDI D+ KTVT++ HP P S+HPC+HA+VMKK++
Sbjct 215 LWLFGYDEQRQPLTVEHMYEDISQDHVKKTVTIENHPHLPPPPMCSVHPCRHAEVMKKII 274
Query 221 DHWRSQGVEPRPDLALIVLLKFISSVVPTI 250
+ G E + L++ LKF+ +V+PTI
Sbjct 275 ETVAEGGGELGVHMYLLIFLKFVQAVIPTI 304
> hsa:64422 ATG3, APG3, APG3-LIKE, APG3L, DKFZp564M1178, FLJ22125,
MGC15201, PC3-96; ATG3 autophagy related 3 homolog (S.
cerevisiae); K08343 autophagy-related protein 3
Length=314
Score = 152 bits (384), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 98/270 (36%), Positives = 142/270 (52%), Gaps = 29/270 (10%)
Query 1 DLLVFKFPTWQWEPAVGKRINSWLPPDKQFLVTRGVACHRRVRDIEASLAAEA---REDA 57
D LV PTWQW ++ ++LP KQFLVT+ V C++R + +E S EA +D
Sbjct 44 DHLVHHCPTWQWATGEELKVKAYLPTGKQFLVTKNVPCYKRCKQMEYSDELEAIIEEDDG 103
Query 58 EG-WVLPVEAEEPEEVLSVSVSGGETNPLEKPPGGSGSSLRDTSGGALPDLKNLRDL--- 113
+G WV + + G T +++ + ++R AL + + D
Sbjct 104 DGGWV---------DTYHNTGITGITEAVKEITLENKDNIRLQDCSALCEEEEDEDEGEA 154
Query 114 -------DSLLCEDDPACADS-STSYFVRNAPDAD----VLSVRTYDLSITYDKYFQTPR 161
+S L E D A D+ + DA +L RTYDL ITYDKY+QTPR
Sbjct 155 ADMEEYEESGLLETDEATLDTRKIVEACKAKTDAGGEDAILQTRTYDLYITYDKYYQTPR 214
Query 162 LWLFGYNESGTPLTPVEIFEDILTDYAAKTVTVDPHPCT-GVPTASIHPCKHAQVMKKVV 220
LWLFGY+E PLT ++EDI D+ KTVT++ HP P S+HPC+HA+VMKK++
Sbjct 215 LWLFGYDEQRQPLTVEHMYEDISQDHVKKTVTIENHPHLPPPPMCSVHPCRHAEVMKKII 274
Query 221 DHWRSQGVEPRPDLALIVLLKFISSVVPTI 250
+ G E + L++ LKF+ +V+PTI
Sbjct 275 ETVAEGGGELGVHMYLLIFLKFVQAVIPTI 304
> cpv:cgd8_2630 APG10/ Aut1p like like autophagocytosis protein
involved in vacuolar transport
Length=242
Score = 146 bits (368), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 84/250 (33%), Positives = 124/250 (49%), Gaps = 66/250 (26%)
Query 1 DLLVFKFPTWQWEPAVGKRINSWLPPDKQFLVTRGVACHRRVRDIEASLAAEAREDAEGW 60
D L+ +FP W W A + I WLP +KQ+L V C +R+ + ++
Sbjct 45 DYLILQFPNWFWRSASEEYIVRWLPQNKQYLHIDNVPCRKRLDSSKLCISK--------- 95
Query 61 VLPVEAEEPEEVLSVSVSGGETNPLEKPPGGSGSSLRDTSGGALPDLKNLRDLDSLLCED 120
N L+ G LP +N+ L ++ D
Sbjct 96 ----------------------NCLDITSDSKGDE------WILPTNENVEKLGNININD 127
Query 121 DPACADSSTSYFVRNAPDADVLSVRTYDLSITYDKYFQTPRLWLFGYNESGTPLTPVEIF 180
D VR YD+S+TYDK+FQTPR+WLFGYN+ G PL+ E+
Sbjct 128 D----------------------VRYYDISVTYDKFFQTPRIWLFGYNKEGYPLSTEEMV 165
Query 181 EDILTDYAAKTVTVDPHPCTGVPTASIHPCKHAQVMKKVVDHWRSQGVEPRPDLALIVLL 240
EDI++DYA KT+T+DPHP TG+ SIHPC H+ ++KK+ + P P L++++LL
Sbjct 166 EDIISDYATKTITLDPHPFTGILCVSIHPCNHSSLLKKMAKN------HP-PHLSIVILL 218
Query 241 KFISSVVPTI 250
KFI++V+P+I
Sbjct 219 KFITTVIPSI 228
> cel:Y55F3AM.4 atg-3; AuTophaGy (yeast Atg homolog) family member
(atg-3); K08343 autophagy-related protein 3
Length=305
Score = 139 bits (349), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 90/265 (33%), Positives = 129/265 (48%), Gaps = 28/265 (10%)
Query 1 DLLVFKFPTWQWEPAVG-KRINSWLPPDKQFLVTRGVACHRRVRDIEASLAAEA---RED 56
D LV PTW+W A +I ++LP DKQFL+TR V CH+R + +E E ED
Sbjct 44 DHLVHHCPTWKWAGASDPSKIRTFLPIDKQFLITRNVPCHKRCKQMEYDEKLEKIINEED 103
Query 57 AE-------GWVLPVEAEEPEEVLSVSVSGGETNPLEKPPGGSGSSLRDTSGGALPDLKN 109
E GWV + + + PP S D D
Sbjct 104 GEYQTSDETGWV-----DTHHYEKEHEKNEEKEQSTAPPPAAPEDSDDDDEEPLDLDELG 158
Query 110 LRDL----DSLLCEDDPACADSSTSYFVRNAPDADVLSVRTYDLSITYDKYFQTPRLWLF 165
L D + + E P A + S +V VRTYDL I YDKY+Q PRL+L
Sbjct 159 LDDDEEDPNRFVVEKKPLAAGNDNS--------GEVEKVRTYDLHICYDKYYQVPRLFLM 210
Query 166 GYNESGTPLTPVEIFEDILTDYAAKTVTVDPHPCTGVPTASIHPCKHAQVMKKVVDHWRS 225
GY+E+ PLT + +ED D++ KT+TV+ HP + ++HPCKHA++MK++++ +
Sbjct 211 GYDENRRPLTVEQTYEDFSADHSNKTITVEAHPSVDLTMPTVHPCKHAEMMKRLINQYAE 270
Query 226 QGVEPRPDLALIVLLKFISSVVPTI 250
G L + LKF+ +V+PTI
Sbjct 271 SGKVLGVHEYLFLFLKFVQAVIPTI 295
> sce:YNR007C ATG3, APG3, AUT1; Atg3p; K08343 autophagy-related
protein 3
Length=310
Score = 119 bits (299), Expect = 8e-27, Method: Compositional matrix adjust.
Identities = 86/283 (30%), Positives = 128/283 (45%), Gaps = 52/283 (18%)
Query 1 DLLVFKFPTWQW-EPAVGKRINSWLPPDKQFLVTRGVACHRRVRDIEASLAAEAREDAEG 59
D L FPTW+W E + +LP +KQFL+ R V C +R
Sbjct 38 DYLCHMFPTWKWNEESSDISYRDFLPKNKQFLIIRKVPCDKRAEQC-------------- 83
Query 60 WVLPVEAEEPEEVLSVSVSGGETNPLEKPPGGSGSSLRDTSGGALPDLKNLRDLDSLLCE 119
VE E P+ ++ G+ + + + G ++ T G D ++ D+D L+ +
Sbjct 84 ----VEVEGPDVIMKGFAEDGDEDDVLEYIGSETEHVQSTPAGGTKD-SSIDDIDELIQD 138
Query 120 DDPACADSSTSYFVRNAPDA---DVLSVRTYDLSITYDKYFQTPRLWLFGYNESGTPLTP 176
+ D + NA D+ R YDL I Y ++ P++++ G+N +G+PL+P
Sbjct 139 MEIKEEDENDDTEEFNAKGGLAKDMAQERYYDLYIAYSTSYRVPKMYIVGFNSNGSPLSP 198
Query 177 VEIFEDILTDYAAKTVTVDPHPC--TGVPTASIHPCKHAQVMKKVVDHWRS--------- 225
++FEDI DY KT T++ P V + SIHPCKHA VMK ++D R
Sbjct 199 EQMFEDISADYRTKTATIEKLPFYKNSVLSVSIHPCKHANVMKILLDKVRVVRQRRRKEL 258
Query 226 ------QGV------------EPRPDLALIVLLKFISSVVPTI 250
GV R D LIV LKFI+SV P+I
Sbjct 259 QEEQELDGVGDWEDLQDDIDDSLRVDQYLIVFLKFITSVTPSI 301
> tpv:TP03_0261 hypothetical protein
Length=197
Score = 87.8 bits (216), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 37/74 (50%), Positives = 51/74 (68%), Gaps = 0/74 (0%)
Query 145 RTYDLSITYDKYFQTPRLWLFGYNESGTPLTPVEIFEDILTDYAAKTVTVDPHPCTGVPT 204
RTY L+ITYD+Y +T R WL GYN+ G PL+ E+FED+ Y KTVTV+ HP TG
Sbjct 117 RTYSLTITYDRYNETARFWLRGYNQFGLPLSKDEMFEDVPEVYVGKTVTVERHPFTGFLN 176
Query 205 ASIHPCKHAQVMKK 218
++HPC +++K+
Sbjct 177 LTVHPCNQKEIVKR 190
> cel:D2085.2 atg-10; AuTophaGy (yeast Atg homolog) family member
(atg-10)
Length=157
Score = 42.7 bits (99), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/95 (24%), Positives = 44/95 (46%), Gaps = 11/95 (11%)
Query 133 VRNAPDADVLSVRTYDLSITYDKYFQTPRLWLFGYNESGTPLTPVEIFEDILTDYAAK-- 190
++ D V++ T+ I Y+ +Q P +W + +G+PL + D+L +
Sbjct 39 LKTLSDGRVITSETH---ILYNSTYQVPTIWFNFFENNGSPLPFRTVIRDVLNISETEES 95
Query 191 ------TVTVDPHPCTGVPTASIHPCKHAQVMKKV 219
++ HP GV +IHPC + +MK++
Sbjct 96 EASIRSRISHYEHPFMGVLYYNIHPCNTSNIMKEL 130
> hsa:83734 ATG10, APG10, APG10L, DKFZp586I0418, FLJ13954, pp12616;
ATG10 autophagy related 10 homolog (S. cerevisiae)
Length=220
Score = 41.6 bits (96), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 25/86 (29%), Positives = 38/86 (44%), Gaps = 7/86 (8%)
Query 144 VRTYDLSITYDKYFQTPRLWLFGYNESGTPLTPVEIFEDILTDYAAK-------TVTVDP 196
V Y+ + Y +Q P L+ G PLT +I+E + Y + T+T
Sbjct 93 VIKYEYHVLYSCSYQVPVLYFRASFLDGRPLTLKDIWEGVHECYKMRLLQGPWDTITQQE 152
Query 197 HPCTGVPTASIHPCKHAQVMKKVVDH 222
HP G P +HPCK + M V+ +
Sbjct 153 HPILGQPFFVLHPCKTNEFMTPVLKN 178
> dre:641330 atg10, MGC123341, im:7153729, zgc:123341; ATG10 autophagy
related 10 homolog (S. cerevisiae)
Length=224
Score = 40.4 bits (93), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 22/92 (23%), Positives = 41/92 (44%), Gaps = 7/92 (7%)
Query 143 SVRTYDLSITYDKYFQTPRLWLFGYNESGTPLTPVEIFEDILTDYAAK-------TVTVD 195
+V Y+ + Y +Q P L+ G L+ E++ ++ +Y + T+T
Sbjct 100 AVLRYEYHVLYSCSYQIPVLYFRASALDGRSLSLEEVWSNVHPNYRQRLKQEPWDTLTQQ 159
Query 196 PHPCTGVPTASIHPCKHAQVMKKVVDHWRSQG 227
HP G P +HPC+ + MK ++ +Q
Sbjct 160 EHPLLGQPFFMLHPCRTEEFMKPALELAHAQN 191
> mmu:66795 Atg10, 5330424L23Rik, 5430428K15Rik, AI852123, APG10,
Agp10, Apg10l, Apg10p, Atg10l; autophagy-related 10 (yeast)
Length=211
Score = 40.0 bits (92), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 25/91 (27%), Positives = 39/91 (42%), Gaps = 7/91 (7%)
Query 139 ADVLSVRTYDLSITYDKYFQTPRLWLFGYNESGTPLTPVEIFEDILTDYAAK-------T 191
A V V ++ + Y +Q P L+ G PL +I+E + Y + T
Sbjct 83 AAVAEVIKHEYHVLYSCSYQVPVLYFRASFLDGRPLALEDIWEGVHECYKPRLLQGPWDT 142
Query 192 VTVDPHPCTGVPTASIHPCKHAQVMKKVVDH 222
+T HP G P +HPCK + M V+ +
Sbjct 143 ITQQEHPILGQPFFVLHPCKTNEFMTAVLKN 173
> ath:AT3G07525 ATG10; ATG10 (autophagy 10); transporter/ ubiquitin
activating enzyme
Length=225
Score = 38.1 bits (87), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 28/108 (25%), Positives = 42/108 (38%), Gaps = 7/108 (6%)
Query 147 YDLSITYDKYFQTPRLWLFGYNESGTPLTPVEIFEDI-------LTDYAAKTVTVDPHPC 199
YD I Y ++ P L+ GY G PL I +D+ L + +T + HP
Sbjct 108 YDFHIVYSASYKVPVLYFRGYCSGGEPLALDVIKKDVPSCSVSLLLESKWTFITQEEHPY 167
Query 200 TGVPTASIHPCKHAQVMKKVVDHWRSQGVEPRPDLALIVLLKFISSVV 247
P +HPC +K + S G + L L+ + VV
Sbjct 168 LNRPWFKLHPCGTEDWIKLLSQSSSSSGCQMPIVLYLVSWFSVVGQVV 215
> ath:AT4G35540 transcription regulator/ zinc ion binding
Length=527
Score = 33.1 bits (74), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 29/119 (24%), Positives = 50/119 (42%), Gaps = 3/119 (2%)
Query 38 CHRRVRDIEASLAAEAREDAEGWVLPVEAEEPEEVLSVSVSGGETNPLEKPPGGSGSSLR 97
C R +++ L A E W V + + + E ++K G+G L
Sbjct 246 CKTRYKELSEKLVKVAEEVGLPWAKDVTVKNVLKHSGTLFALMEAKSMKKRKQGTGKELV 305
Query 98 DTSGGALPDL-KNLRDLDSLLCEDDPACADSSTSYFVRNAPDADVLSVRTYDLSITYDK 155
T G + DL + +S+ C DD A D+ + YF + LS+ YD +I+ ++
Sbjct 306 RTDGFCVEDLVMDCLSKESMYCYDDDARQDTMSRYF--DVEGERQLSLCNYDDNISENQ 362
> eco:b3765 yifB, ECK3758, JW3738; predicted bifunctional enzyme
and transcriptional regulator; K07391 magnesium chelatase
family protein
Length=506
Score = 30.4 bits (67), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 25/52 (48%), Gaps = 0/52 (0%)
Query 64 VEAEEPEEVLSVSVSGGETNPLEKPPGGSGSSLRDTSGGALPDLKNLRDLDS 115
+ E+ + L ++ +GG L PPG + L G LPDL N L+S
Sbjct 194 IGQEQGKRGLEITAAGGHNLLLIGPPGTGKTMLASRINGLLPDLSNEEALES 245
Lambda K H
0.317 0.135 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8956755920
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40