bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1106_orf1
Length=132
Score E
Sequences producing significant alignments: (Bits) Value
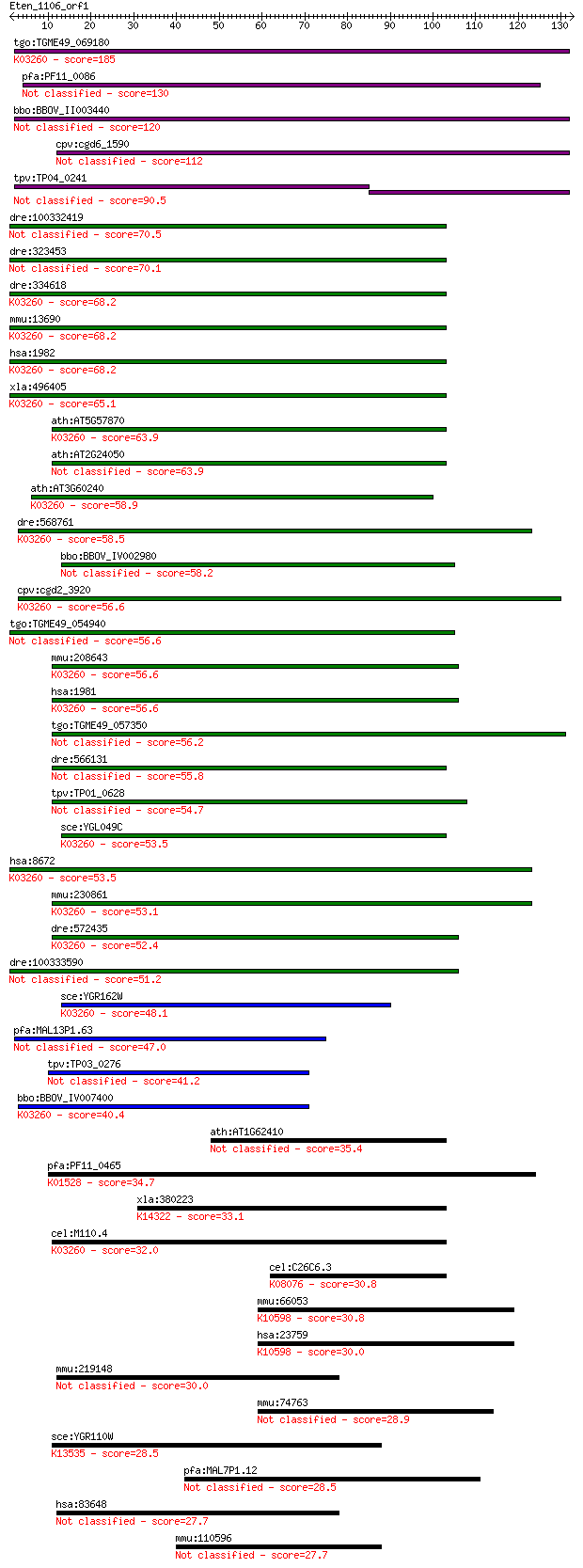
tgo:TGME49_069180 MIF4G domain-containing protein (EC:3.1.1.32... 185 3e-47
pfa:PF11_0086 MIF4G domain containing protein 130 9e-31
bbo:BBOV_II003440 18.m06288; MIF4G domain containing protein 120 1e-27
cpv:cgd6_1590 eukaryotic translation initiation factor 4 gamma... 112 3e-25
tpv:TP04_0241 hypothetical protein 90.5 1e-18
dre:100332419 eukaryotic translation initiation factor 4, gamm... 70.5 1e-12
dre:323453 eif4g2a, MGC192940, NAT1A, im:7148615, wu:fb81f07, ... 70.1 2e-12
dre:334618 eif4g2b, NAT1B, eif4g2, hm:zeh1307, wu:fa14h01, wu:... 68.2 6e-12
mmu:13690 Eif4g2, AA589388, DAP-5, E130105L11Rik, Nat1, Natm1,... 68.2 7e-12
hsa:1982 EIF4G2, AAG1, DAP5, FLJ41344, NAT1, P97; eukaryotic t... 68.2 7e-12
xla:496405 nat1, DAP5, EIF4G2, p97; eIF4G-related protein NAT1... 65.1 5e-11
ath:AT5G57870 eukaryotic translation initiation factor 4F, put... 63.9 1e-10
ath:AT2G24050 MIF4G domain-containing protein / MA3 domain-con... 63.9 1e-10
ath:AT3G60240 EIF4G; EIF4G (EUKARYOTIC TRANSLATION INITIATION ... 58.9 4e-09
dre:568761 si:dkey-1a7.2; K03260 translation initiation factor 4G 58.5 5e-09
bbo:BBOV_IV002980 21.m02788; eukaryotic initiation factor 4G m... 58.2 6e-09
cpv:cgd2_3920 eIF4G eukaryotic initiation factor 4, Nic domain... 56.6 2e-08
tgo:TGME49_054940 hypothetical protein 56.6 2e-08
mmu:208643 Eif4g1, E030015G23Rik, MGC37551, MGC90776, eIF4GI; ... 56.6 2e-08
hsa:1981 EIF4G1, DKFZp686A1451, EIF-4G1, EIF4F, EIF4G, EIF4GI,... 56.6 2e-08
tgo:TGME49_057350 eukaryotic translation initiation factor, pu... 56.2 3e-08
dre:566131 eukaryotic translation initiation factor 4 gamma, 3... 55.8 3e-08
tpv:TP01_0628 hypothetical protein 54.7 8e-08
sce:YGL049C Tif4632p; K03260 translation initiation factor 4G 53.5 2e-07
hsa:8672 EIF4G3, eIF-4G_3, eIF4G_3, eIF4GII; eukaryotic transl... 53.5 2e-07
mmu:230861 Eif4g3, 1500002J22Rik, 4833436O05, 4930523M17Rik, G... 53.1 2e-07
dre:572435 MGC158450, wu:fb50a05, wu:fc60b06; zgc:158450; K032... 52.4 3e-07
dre:100333590 eukaryotic translation initiation factor 4 gamma... 51.2 7e-07
sce:YGR162W Tif4631p; K03260 translation initiation factor 4G 48.1 7e-06
pfa:MAL13P1.63 asparagine-rich protein 47.0 2e-05
tpv:TP03_0276 hypothetical protein 41.2 0.001
bbo:BBOV_IV007400 23.m06334; MIF4G domain containing protein; ... 40.4 0.002
ath:AT1G62410 MIF4G domain-containing protein 35.4 0.050
pfa:PF11_0465 dyn1; dynamin-like protein; K01528 dynamin GTPas... 34.7 0.079
xla:380223 paip1, MGC154295, MGC53704; poly(A) binding protein... 33.1 0.25
cel:M110.4 ifg-1; Initiation Factor 4G (eIF4G) family member (... 32.0 0.50
cel:C26C6.3 nas-36; Nematode AStacin protease family member (n... 30.8 1.0
mmu:66053 Ppil2, 0610009L05Rik, 1700016N17Rik, 4921520K19Rik, ... 30.8 1.0
hsa:23759 PPIL2, CYC4, CYP60, Cyp-60, FLJ39930, MGC33174, MGC7... 30.0 2.0
mmu:219148 Fam167a, A030013D21; family with sequence similarit... 30.0 2.1
mmu:74763 Nat15, 1200013P24Rik, AI315146; N-acetyltransferase ... 28.9 3.9
sce:YGR110W CLD1; Cld1p; K13535 cardiolipin-specific phospholi... 28.5 5.3
pfa:MAL7P1.12 erythrocyte membrane-associated antigen 28.5 6.3
hsa:83648 FAM167A, C8orf13, D8S265, DKFZp761G151, MGC120649, M... 27.7 8.8
mmu:110596 Rgnef, 9230110L08Rik, AI323540, D13Bwg1089e, RIP2, ... 27.7 10.0
> tgo:TGME49_069180 MIF4G domain-containing protein (EC:3.1.1.32
3.4.21.72); K03260 translation initiation factor 4G
Length=2668
Score = 185 bits (469), Expect = 3e-47, Method: Composition-based stats.
Identities = 83/130 (63%), Positives = 107/130 (82%), Gaps = 0/130 (0%)
Query 2 KQKEDAQMVLLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQHH 61
+QK+D Q LLR LKG LNKLT+E+F+++Y QILNAGIK+++EV ALM+M+F+KAVSQHH
Sbjct 2083 EQKKDEQQQLLRRLKGHLNKLTLEKFEKLYPQILNAGIKEKEEVNALMKMVFEKAVSQHH 2142
Query 62 FIQMYVMLCGKLKTDFQELLQDDHRSTKFRRILIDQCEDSFNANLEPIAVPQGLSEDDAY 121
FIQMYV LC +LK D +++L DD + + FRRILI+QCEDSF ANLEP+ VP+GL ED+A+
Sbjct 2143 FIQMYVQLCSRLKDDLKQILADDAKGSYFRRILINQCEDSFVANLEPMRVPEGLDEDEAF 2202
Query 122 EFELNYKKTM 131
EF YK M
Sbjct 2203 EFAQLYKARM 2212
> pfa:PF11_0086 MIF4G domain containing protein
Length=3334
Score = 130 bits (328), Expect = 9e-31, Method: Compositional matrix adjust.
Identities = 58/121 (47%), Positives = 87/121 (71%), Gaps = 0/121 (0%)
Query 4 KEDAQMVLLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQHHFI 63
K D ++R L+G+LNKLT E+FD +Y+QI+ GI + E+ LM+++F+KAV+QHHF+
Sbjct 2681 KADKYTCMMRKLRGILNKLTFEKFDLLYEQIILVGITKLDEIIGLMKLVFEKAVTQHHFV 2740
Query 64 QMYVMLCGKLKTDFQELLQDDHRSTKFRRILIDQCEDSFNANLEPIAVPQGLSEDDAYEF 123
QMYV LC KL F + +D + +F++ILI+QC+DSF NL+PI P L+E++ +EF
Sbjct 2741 QMYVQLCKKLNVHFHNMKLEDDTTVQFKKILINQCQDSFENNLKPIEFPSHLNEEEKFEF 2800
Query 124 E 124
E
Sbjct 2801 E 2801
> bbo:BBOV_II003440 18.m06288; MIF4G domain containing protein
Length=1043
Score = 120 bits (300), Expect = 1e-27, Method: Composition-based stats.
Identities = 60/149 (40%), Positives = 88/149 (59%), Gaps = 19/149 (12%)
Query 2 KQKEDAQMVLLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQHH 61
K K D + L+R L GLLN+LT+E+FD IY+Q+L++G++ + L+++IF+KA++QHH
Sbjct 569 KLKMDKEQQLIRKLMGLLNRLTVEKFDTIYKQVLSSGVETYNQAFVLVKIIFEKAITQHH 628
Query 62 FIQMYVMLCGKLKTDFQELLQDDH-------------------RSTKFRRILIDQCEDSF 102
FI MYV LC KL D ++ R + F +IL++ C+DSF
Sbjct 629 FIPMYVELCAKLSYDLNNFTDNEDASVAQGDSSTTGKSDQKATRRSDFMKILLNCCQDSF 688
Query 103 NANLEPIAVPQGLSEDDAYEFELNYKKTM 131
NL+P+ P L DD +EFEL YK M
Sbjct 689 ENNLKPMECPPDLEGDDKFEFELKYKHRM 717
> cpv:cgd6_1590 eukaryotic translation initiation factor 4 gamma;
Nic domain containing protein
Length=1184
Score = 112 bits (280), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 53/122 (43%), Positives = 86/122 (70%), Gaps = 7/122 (5%)
Query 12 LRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQHHFIQMYVMLCG 71
LR + +LNKLTIE+FD++Y+QIL+ GI E+E+ L++++FDKA +QHHFI MYV LC
Sbjct 702 LRQYRAILNKLTIEKFDKLYEQILSVGINNEEEMIGLLKLVFDKATTQHHFIPMYVELCD 761
Query 72 KLKTDFQELLQDDHRSTKFRRILIDQCEDSFNANLEPIAVPQGL--SEDDAYEFELNYKK 129
KL+ +++ + + RRIL+D C++ F NL + +P+ + +E+D++E++L YK
Sbjct 762 KLRDHLKDVT-----TIETRRILVDLCQELFVENLSEMVLPEHIQDNEEDSFEWQLKYKN 816
Query 130 TM 131
M
Sbjct 817 KM 818
> tpv:TP04_0241 hypothetical protein
Length=1429
Score = 90.5 bits (223), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 42/83 (50%), Positives = 56/83 (67%), Gaps = 0/83 (0%)
Query 2 KQKEDAQMVLLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQHH 61
KQK D +M+L R + GLLN+LT E+FD IY QI+ GI + + L++ +FDKAV+QHH
Sbjct 651 KQKNDKEMLLRRKIMGLLNRLTFEKFDIIYNQIIECGIDTPEHAEMLVKFVFDKAVTQHH 710
Query 62 FIQMYVMLCGKLKTDFQELLQDD 84
FI MYV LC KL D + Q +
Sbjct 711 FIPMYVELCAKLAIDLYTIDQSN 733
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 20/47 (42%), Positives = 30/47 (63%), Gaps = 0/47 (0%)
Query 85 HRSTKFRRILIDQCEDSFNANLEPIAVPQGLSEDDAYEFELNYKKTM 131
+R + F RIL++ +DSF NL+P+ +P L DD +E+E YK M
Sbjct 790 NRKSDFMRILLNCSQDSFEDNLKPLEIPAELEGDDRFEYEQKYKHKM 836
> dre:100332419 eukaryotic translation initiation factor 4, gamma
2a-like
Length=953
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 37/109 (33%), Positives = 64/109 (58%), Gaps = 10/109 (9%)
Query 1 EKQKEDAQMVLLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQH 60
EK++ DA + R ++G+LNKLT E+FD++ ++LN G+ + +K ++ +I DKA+ +
Sbjct 137 EKERHDA---IFRKVRGILNKLTPEKFDKLCLELLNVGVDSKLVLKGIILLIVDKALEEP 193
Query 61 HFIQMYVMLCGKLKTDFQ-------ELLQDDHRSTKFRRILIDQCEDSF 102
+ +Y LC +L D E+ +ST FRR+LI + +D F
Sbjct 194 KYSSLYAQLCLRLAEDAPNFDGPSTEIQSSQKQSTTFRRLLISKLQDEF 242
> dre:323453 eif4g2a, MGC192940, NAT1A, im:7148615, wu:fb81f07,
wu:fb82b06, wu:fb98h08; eukaryotic translation initiation
factor 4, gamma 2a
Length=891
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 37/109 (33%), Positives = 64/109 (58%), Gaps = 10/109 (9%)
Query 1 EKQKEDAQMVLLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQH 60
EK++ DA + R ++G+LNKLT E+FD++ ++LN G+ + +K ++ +I DKA+ +
Sbjct 65 EKERHDA---IFRKVRGILNKLTPEKFDKLCLELLNVGVDSKLVLKGIILLIVDKALEEP 121
Query 61 HFIQMYVMLCGKLKTDFQ-------ELLQDDHRSTKFRRILIDQCEDSF 102
+ +Y LC +L D E+ +ST FRR+LI + +D F
Sbjct 122 KYSSLYAQLCLRLAEDAPNFDGPSTEIQSSQKQSTTFRRLLISKLQDEF 170
> dre:334618 eif4g2b, NAT1B, eif4g2, hm:zeh1307, wu:fa14h01, wu:fb44h01,
wu:fb59c06, wu:fc21b05; eukaryotic translation initiation
factor 4, gamma 2b; K03260 translation initiation factor
4G
Length=898
Score = 68.2 bits (165), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 37/109 (33%), Positives = 63/109 (57%), Gaps = 10/109 (9%)
Query 1 EKQKEDAQMVLLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQH 60
EK+ DA + R ++G+LNKLT E+FD++ ++LN G+ + +K ++ +I DKA+ +
Sbjct 65 EKEHHDA---IFRKVRGILNKLTPEKFDKLCLELLNVGVDSKLVLKGIILLIVDKALEEP 121
Query 61 HFIQMYVMLCGKLKTD-------FQELLQDDHRSTKFRRILIDQCEDSF 102
+ +Y LC +L D E+ +ST FRR+LI + +D F
Sbjct 122 KYSSLYAQLCLRLAEDAPNFDGPTPEIQSSQKQSTTFRRLLITKLQDEF 170
> mmu:13690 Eif4g2, AA589388, DAP-5, E130105L11Rik, Nat1, Natm1,
p97; eukaryotic translation initiation factor 4, gamma 2;
K03260 translation initiation factor 4G
Length=868
Score = 68.2 bits (165), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 37/109 (33%), Positives = 64/109 (58%), Gaps = 10/109 (9%)
Query 1 EKQKEDAQMVLLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQH 60
EK++ DA + R ++G+LNKLT E+FD++ ++LN G++ + +K ++ +I DKA+ +
Sbjct 70 EKERHDA---IFRKVRGILNKLTPEKFDKLCLELLNVGVESKLILKGVILLIVDKALEEP 126
Query 61 HFIQMYVMLCGKLKTDFQ-------ELLQDDHRSTKFRRILIDQCEDSF 102
+ +Y LC +L D E +ST FRR+LI + +D F
Sbjct 127 KYSSLYAQLCLRLAEDAPNFDGPAAEGQPGQKQSTTFRRLLISKLQDEF 175
> hsa:1982 EIF4G2, AAG1, DAP5, FLJ41344, NAT1, P97; eukaryotic
translation initiation factor 4 gamma, 2; K03260 translation
initiation factor 4G
Length=869
Score = 68.2 bits (165), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 37/109 (33%), Positives = 64/109 (58%), Gaps = 10/109 (9%)
Query 1 EKQKEDAQMVLLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQH 60
EK++ DA + R ++G+LNKLT E+FD++ ++LN G++ + +K ++ +I DKA+ +
Sbjct 70 EKERHDA---IFRKVRGILNKLTPEKFDKLCLELLNVGVESKLILKGVILLIVDKALEEP 126
Query 61 HFIQMYVMLCGKLKTDFQ-------ELLQDDHRSTKFRRILIDQCEDSF 102
+ +Y LC +L D E +ST FRR+LI + +D F
Sbjct 127 KYSSLYAQLCLRLAEDAPNFDGPAAEGQPGQKQSTTFRRLLISKLQDEF 175
> xla:496405 nat1, DAP5, EIF4G2, p97; eIF4G-related protein NAT1;
K03260 translation initiation factor 4G
Length=903
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 35/109 (32%), Positives = 61/109 (55%), Gaps = 10/109 (9%)
Query 1 EKQKEDAQMVLLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQH 60
EK++ DA + R ++G+LNKL E+FD++ ++LN G+ + +K ++ +I DKA+ +
Sbjct 75 EKERHDA---IFRKVRGILNKLAPEKFDKLCLELLNVGVDSKLILKGVILLIVDKALEEP 131
Query 61 HFIQMYVMLCGKLKTDFQEL-------LQDDHRSTKFRRILIDQCEDSF 102
+ +Y LC +L D +ST FRR+LI + +D F
Sbjct 132 KYSSLYAQLCLRLAEDAPNFDGPSADGPPGQKQSTTFRRLLISKLQDEF 180
> ath:AT5G57870 eukaryotic translation initiation factor 4F, putative
/ eIF-4F, putative; K03260 translation initiation factor
4G
Length=776
Score = 63.9 bits (154), Expect = 1e-10, Method: Composition-based stats.
Identities = 30/94 (31%), Positives = 55/94 (58%), Gaps = 2/94 (2%)
Query 11 LLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQHHFIQMYVMLC 70
+L+ +KG+LNKLT E++D + Q++ +GI +K ++ +IFDKAV + F MY LC
Sbjct 211 VLKTVKGILNKLTPEKYDLLKGQLIESGITSADILKGVITLIFDKAVLEPTFCPMYAKLC 270
Query 71 GKLKTDFQEL--LQDDHRSTKFRRILIDQCEDSF 102
+ + + F+R+L++ C+++F
Sbjct 271 SDINDQLPTFPPAEPGDKEITFKRVLLNICQEAF 304
> ath:AT2G24050 MIF4G domain-containing protein / MA3 domain-containing
protein
Length=747
Score = 63.9 bits (154), Expect = 1e-10, Method: Composition-based stats.
Identities = 28/94 (29%), Positives = 59/94 (62%), Gaps = 2/94 (2%)
Query 11 LLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQHHFIQMYVMLC 70
+++ +KG+LNKLT E+++ + Q+++AGI +K ++Q+IF+ A+ Q F +MY +LC
Sbjct 174 VVKSVKGILNKLTPEKYELLKGQLIDAGITSADILKEVIQLIFENAILQPTFCEMYALLC 233
Query 71 GKLKTDFQELLQDD--HRSTKFRRILIDQCEDSF 102
+ ++ + F+R+L++ C+++F
Sbjct 234 FDINGQLPSFPSEEPGGKEITFKRVLLNNCQEAF 267
> ath:AT3G60240 EIF4G; EIF4G (EUKARYOTIC TRANSLATION INITIATION
FACTOR 4G); translation initiation factor; K03260 translation
initiation factor 4G
Length=1723
Score = 58.9 bits (141), Expect = 4e-09, Method: Composition-based stats.
Identities = 27/94 (28%), Positives = 53/94 (56%), Gaps = 1/94 (1%)
Query 6 DAQMVLLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQHHFIQM 65
D + R LK +LNKLT + F+++++Q+ + I + ++ IFDKA+ + F +M
Sbjct 1086 DEEQAKQRQLKSILNKLTPQNFEKLFEQVKSVNIDNAVTLSGVISQIFDKALMEPTFCEM 1145
Query 66 YVMLCGKLKTDFQELLQDDHRSTKFRRILIDQCE 99
Y C L + ++ + T F+R+L+++C+
Sbjct 1146 YADFCFHLSGALPDFNENGEKIT-FKRLLLNKCQ 1178
> dre:568761 si:dkey-1a7.2; K03260 translation initiation factor
4G
Length=1535
Score = 58.5 bits (140), Expect = 5e-09, Method: Composition-based stats.
Identities = 32/122 (26%), Positives = 65/122 (53%), Gaps = 11/122 (9%)
Query 3 QKEDAQMV--LLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQH 60
+ DAQ L R ++ +LNKLT + F ++ +Q+ + I E+ +K ++ ++F+KA+++
Sbjct 694 EDPDAQRTQELFRKVRSILNKLTPQMFSQLMKQVTDLTIDTEERLKGVIDLVFEKAINEP 753
Query 61 HFIQMYVMLCGKLKTDFQELLQDDHRSTKFRRILIDQCEDSFNANLEPIAVPQGLSEDDA 120
F Y +C L T + + + FR++L+++C+ F + +DDA
Sbjct 754 SFSVAYGNMCSCLATLKVPMTDKPNSTVNFRKLLLNRCQKEFEKD---------KMDDDA 804
Query 121 YE 122
+E
Sbjct 805 FE 806
> bbo:BBOV_IV002980 21.m02788; eukaryotic initiation factor 4G
middle domain containing protein
Length=1481
Score = 58.2 bits (139), Expect = 6e-09, Method: Composition-based stats.
Identities = 30/94 (31%), Positives = 56/94 (59%), Gaps = 2/94 (2%)
Query 13 RHLKGLLNKLTIERFDRIYQQ--ILNAGIKQEKEVKALMQMIFDKAVSQHHFIQMYVMLC 70
R ++GLLNKLT+E+F + + +L + E++V A+ +I DK+VS+ + +Y L
Sbjct 930 RTVRGLLNKLTVEKFLTVAENLAVLYEKLTHEEDVDAMADLIIDKSVSELDYSDVYADLA 989
Query 71 GKLKTDFQELLQDDHRSTKFRRILIDQCEDSFNA 104
+ F + ++T F R+L+++ +DSF+A
Sbjct 990 YLISCRFNDNFDVGSKNTLFYRVLLNKLQDSFDA 1023
> cpv:cgd2_3920 eIF4G eukaryotic initiation factor 4, Nic domain
containing protein ; K03260 translation initiation factor
4G
Length=1235
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 41/133 (30%), Positives = 70/133 (52%), Gaps = 8/133 (6%)
Query 3 QKEDAQMVLLRHLKGLLNKLTIERFDRIYQQILNA--GIKQEKEVKALMQMIFDKAVSQH 60
Q D+ + R +K LLNK+T+E+F I +++ IK E++ L++ + DKA+++
Sbjct 929 QTVDSLETVRRAVKSLLNKITVEKFTVIAEKLAVCIDEIKNVDELEELVKQVLDKAITEP 988
Query 61 HFIQMYVMLCGKLKTDFQELLQDDHRST-KFRRILIDQCEDSFN---ANLEPIAVPQGLS 116
F +MY LC LK L+ D +ST F + L+ +CE F N+ P +
Sbjct 989 DFSEMYADLCQILKWRSPVLVNKDGKSTIAFSKALLARCEQEFKNMPRNMTP--TDEERE 1046
Query 117 EDDAYEFELNYKK 129
+ D+ E + Y+K
Sbjct 1047 KYDSEELAILYRK 1059
> tgo:TGME49_054940 hypothetical protein
Length=3799
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 31/109 (28%), Positives = 61/109 (55%), Gaps = 7/109 (6%)
Query 1 EKQKEDAQMVLLRHLKGLLNKLTIERFDRIYQQILNAG--IKQEKEVKALMQMIFDKAVS 58
+KQ D +L R + LLNK++IE+F + ++I G ++ +E++ L +++ KAV+
Sbjct 3092 KKQTLDRPQLLRRSINSLLNKMSIEKFVVVAEKIAVEGESLENAEELQMLADLVYAKAVT 3151
Query 59 QHHFIQMYVMLCGKLK---TDFQELLQDDHRSTKFRRILIDQCEDSFNA 104
+ + + Y LC LK DF + + + + F R +++C++ F A
Sbjct 3152 EPEYSETYADLCQLLKWRSLDFDK--EGEEKQLNFNRAFVNRCQEEFEA 3198
> mmu:208643 Eif4g1, E030015G23Rik, MGC37551, MGC90776, eIF4GI;
eukaryotic translation initiation factor 4, gamma 1; K03260
translation initiation factor 4G
Length=1593
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 26/95 (27%), Positives = 53/95 (55%), Gaps = 0/95 (0%)
Query 11 LLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQHHFIQMYVMLC 70
L R ++ +LNKLT + F ++ +Q+ I E+ +K ++ +IF+KA+S+ +F Y +C
Sbjct 757 LFRRVRSILNKLTPQMFQQLMKQVTQLAIDTEERLKGVIDLIFEKAISEPNFSVAYANMC 816
Query 71 GKLKTDFQELLQDDHRSTKFRRILIDQCEDSFNAN 105
L + + FR++L+++C+ F +
Sbjct 817 RCLMALKVPTTEKPTVTVNFRKLLLNRCQKEFEKD 851
> hsa:1981 EIF4G1, DKFZp686A1451, EIF-4G1, EIF4F, EIF4G, EIF4GI,
P220; eukaryotic translation initiation factor 4 gamma, 1;
K03260 translation initiation factor 4G
Length=1606
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 26/95 (27%), Positives = 53/95 (55%), Gaps = 0/95 (0%)
Query 11 LLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQHHFIQMYVMLC 70
L R ++ +LNKLT + F ++ +Q+ I E+ +K ++ +IF+KA+S+ +F Y +C
Sbjct 767 LFRRVRSILNKLTPQMFQQLMKQVTQLAIDTEERLKGVIDLIFEKAISEPNFSVAYANMC 826
Query 71 GKLKTDFQELLQDDHRSTKFRRILIDQCEDSFNAN 105
L + + FR++L+++C+ F +
Sbjct 827 RCLMALKVPTTEKPTVTVNFRKLLLNRCQKEFEKD 861
> tgo:TGME49_057350 eukaryotic translation initiation factor,
putative
Length=820
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 38/132 (28%), Positives = 73/132 (55%), Gaps = 14/132 (10%)
Query 11 LLRHLKGLLNKLTIERFDRIYQQILNA--GIKQEKEVKALMQMIFDKAVSQHHFIQMYVM 68
L R +K LLNKLTIE+FD I +++ G+K+ KE++ ++ ++ +KAV++ + +MY
Sbjct 498 LDRTVKALLNKLTIEKFDTITEKLARTVEGLKEHKELQLVVNVVMEKAVTEPDWSEMYAD 557
Query 69 LCGKLK-----TDFQELLQDDHRSTKFRRILIDQCEDSFNANLEPI-----AVPQGLSED 118
LC L+ T+ ++ ++ R T F + L+ + + + A + ++P G +
Sbjct 558 LCQVLQWRSVPTEGTDI--ENFRKTPFMKALLTKIQAEYEAMPRSLESRLRSLPSGELDP 615
Query 119 DAYEFELNYKKT 130
+A E K+T
Sbjct 616 EAIEEMQQLKRT 627
> dre:566131 eukaryotic translation initiation factor 4 gamma,
3-like
Length=1697
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 25/92 (27%), Positives = 52/92 (56%), Gaps = 0/92 (0%)
Query 11 LLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQHHFIQMYVMLC 70
L R ++ +LNKLT + F ++ +Q+ + I E+ +K ++ ++F+KA+ + F Y +C
Sbjct 864 LFRKVRSILNKLTPQMFHQLMKQVTDLTINTEERLKGVIDLVFEKAIDEPSFSVAYANMC 923
Query 71 GKLKTDFQELLQDDHRSTKFRRILIDQCEDSF 102
L T + + FR++L+++C+ F
Sbjct 924 RCLATLKVPTADKPNTTVNFRKLLLNRCQKEF 955
> tpv:TP01_0628 hypothetical protein
Length=1134
Score = 54.7 bits (130), Expect = 8e-08, Method: Composition-based stats.
Identities = 32/106 (30%), Positives = 63/106 (59%), Gaps = 11/106 (10%)
Query 11 LLRHLKGLLNKLTIERFDRIYQQILNAGIKQE----KEVKALMQMIFDKAVSQHHFIQMY 66
L R ++ LLNKLT+E F + ++I+ I +E +EV+ L+ ++ +K ++ + MY
Sbjct 739 LKRSIRTLLNKLTVENFLVVSEKIV--AIYREMSLREEVEVLVDLLHEKGTTELEYADMY 796
Query 67 VMLCGKLKTDFQELLQDDHRSTKFRRILIDQCEDSF-----NANLE 107
L L+ + ++L +++T F + L+++C+DSF NA+L+
Sbjct 797 ADLAFLLRYSYNDILDLGNKTTLFHKSLLNKCQDSFEQINSNASLD 842
> sce:YGL049C Tif4632p; K03260 translation initiation factor 4G
Length=914
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 33/100 (33%), Positives = 51/100 (51%), Gaps = 10/100 (10%)
Query 13 RHLKGLLNKLTIERFDRIYQQILNAGIKQEKE-----VKALMQMIFDKAVSQHHFIQMYV 67
R +K LLNKLT+E FD I +IL+ + + E +K +++ IF KA + H+ MY
Sbjct 568 RKMKSLLNKLTLEMFDSISSEILDIANQSKWEDDGETLKIVIEQIFHKACDEPHWSSMYA 627
Query 68 MLCGKLKTDFQELLQDDHRSTK-----FRRILIDQCEDSF 102
LCGK+ D ++D K L+ +C + F
Sbjct 628 QLCGKVVKDLDPNIKDKENEGKNGPKLVLHYLVARCHEEF 667
> hsa:8672 EIF4G3, eIF-4G_3, eIF4G_3, eIF4GII; eukaryotic translation
initiation factor 4 gamma, 3; K03260 translation initiation
factor 4G
Length=1621
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 29/126 (23%), Positives = 65/126 (51%), Gaps = 13/126 (10%)
Query 1 EKQKEDAQMV----LLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKA 56
+ Q +D + + L R ++ +LNKLT + F+++ +Q+ + E+ +K ++ ++F+KA
Sbjct 776 DSQADDPENIKTQELFRKVRSILNKLTPQMFNQLMKQVSGLTVDTEERLKGVIDLVFEKA 835
Query 57 VSQHHFIQMYVMLCGKLKTDFQELLQDDHRSTKFRRILIDQCEDSFNANLEPIAVPQGLS 116
+ + F Y +C L T + + FR++L+++C+ F + +
Sbjct 836 IDEPSFSVAYANMCRCLVTLKVPMADKPGNTVNFRKLLLNRCQKEFEKD---------KA 886
Query 117 EDDAYE 122
+DD +E
Sbjct 887 DDDVFE 892
> mmu:230861 Eif4g3, 1500002J22Rik, 4833436O05, 4930523M17Rik,
G1-419-52, eIF4GII, repro8; eukaryotic translation initiation
factor 4 gamma, 3; K03260 translation initiation factor 4G
Length=1578
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 27/112 (24%), Positives = 59/112 (52%), Gaps = 9/112 (8%)
Query 11 LLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQHHFIQMYVMLC 70
L R ++ +LNKLT + F+++ +Q+ + E+ +K ++ ++F+KA+ + F Y +C
Sbjct 767 LFRKVRSILNKLTPQMFNQLMKQVSALTVDTEERLKGVIDLVFEKAIDEPSFSVAYANMC 826
Query 71 GKLKTDFQELLQDDHRSTKFRRILIDQCEDSFNANLEPIAVPQGLSEDDAYE 122
L T + + FR++L+++C+ F + ++DD +E
Sbjct 827 RCLVTLKVPMADKPGNTVNFRKLLLNRCQKEFEKD---------KADDDVFE 869
> dre:572435 MGC158450, wu:fb50a05, wu:fc60b06; zgc:158450; K03260
translation initiation factor 4G
Length=1585
Score = 52.4 bits (124), Expect = 3e-07, Method: Composition-based stats.
Identities = 26/95 (27%), Positives = 52/95 (54%), Gaps = 0/95 (0%)
Query 11 LLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQHHFIQMYVMLC 70
L R ++ +LNKLT + F ++ +Q+ I E+ +K ++ +IF+KA+S+ +F Y +C
Sbjct 728 LFRRVRSVLNKLTPQMFQQLMKQVTELAIDTEERLKGVIDLIFEKAISEPNFSVAYANMC 787
Query 71 GKLKTDFQELLQDDHRSTKFRRILIDQCEDSFNAN 105
L + FR++L+++C+ F +
Sbjct 788 RCLMGLKVPTSDKPGVTVNFRKLLLNRCQKEFEKD 822
> dre:100333590 eukaryotic translation initiation factor 4 gamma,
1-like
Length=966
Score = 51.2 bits (121), Expect = 7e-07, Method: Composition-based stats.
Identities = 28/105 (26%), Positives = 53/105 (50%), Gaps = 0/105 (0%)
Query 1 EKQKEDAQMVLLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQH 60
E Q E L + ++ +LNKLT + F + +Q+ I E+ +K ++ +IF+KA+S+
Sbjct 552 EDQPELQTQELFKRVRSILNKLTPQMFQPLMKQVTELSIHTEERLKGVIDLIFEKAISEP 611
Query 61 HFIQMYVMLCGKLKTDFQELLQDDHRSTKFRRILIDQCEDSFNAN 105
+F Y +C L FR++L+++C+ F +
Sbjct 612 NFSVAYANMCRCLMGLKVPTADKPGVFVNFRKLLLNRCQKEFEKD 656
> sce:YGR162W Tif4631p; K03260 translation initiation factor 4G
Length=952
Score = 48.1 bits (113), Expect = 7e-06, Method: Composition-based stats.
Identities = 30/82 (36%), Positives = 46/82 (56%), Gaps = 5/82 (6%)
Query 13 RHLKGLLNKLTIERFDRIYQQIL---NAGIKQE--KEVKALMQMIFDKAVSQHHFIQMYV 67
R +K LLNKLT+E FD I +IL N + + + +KA+++ IF KA + H+ MY
Sbjct 608 RKMKSLLNKLTLEMFDAISSEILAIANISVWETNGETLKAVIEQIFLKACDEPHWSSMYA 667
Query 68 MLCGKLKTDFQELLQDDHRSTK 89
LCGK+ + + D+ K
Sbjct 668 QLCGKVVKELNPDITDETNEGK 689
> pfa:MAL13P1.63 asparagine-rich protein
Length=1255
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 28/78 (35%), Positives = 47/78 (60%), Gaps = 6/78 (7%)
Query 2 KQKEDAQMV--LLRHLKGLLNKLTIERFDRIYQ---QILNAGIKQEKEVKALMQMIFDKA 56
K+KE+ + + R +K LLNKLT+E F I + QI+ + + + E++ ++ + DKA
Sbjct 950 KKKENMSPIEYMERQVKSLLNKLTVENFPIITEKMCQIMESRLNTD-EIQTVVNQVIDKA 1008
Query 57 VSQHHFIQMYVMLCGKLK 74
V +H + +MY LC LK
Sbjct 1009 VLEHDWSEMYADLCQTLK 1026
> tpv:TP03_0276 hypothetical protein
Length=359
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 39/63 (61%), Gaps = 2/63 (3%)
Query 10 VLLRHLKGLLNKLTIERFDRIYQQIL--NAGIKQEKEVKALMQMIFDKAVSQHHFIQMYV 67
+ R ++G LNKLTIE+F+ + ++I + +E++ ++ +I DKAV + + +MY
Sbjct 68 ITCRKIQGNLNKLTIEKFETLAEKIAIQCESLSNYEELQKIVDIIIDKAVLEPDWSEMYS 127
Query 68 MLC 70
LC
Sbjct 128 DLC 130
> bbo:BBOV_IV007400 23.m06334; MIF4G domain containing protein;
K03260 translation initiation factor 4G
Length=367
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 40/70 (57%), Gaps = 2/70 (2%)
Query 3 QKEDAQMVLLRHLKGLLNKLTIERFDRIYQQIL--NAGIKQEKEVKALMQMIFDKAVSQH 60
++ A L R LK LNKLTIE+F+ + ++I ++Q ++ + +I DKA+ +
Sbjct 66 SRQGAGDPLTRQLKSNLNKLTIEKFETLAEKIAIQCESLEQYDQLCVAVDLILDKALQEP 125
Query 61 HFIQMYVMLC 70
+ +MY LC
Sbjct 126 EWSEMYADLC 135
> ath:AT1G62410 MIF4G domain-containing protein
Length=223
Score = 35.4 bits (80), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 28/57 (49%), Gaps = 2/57 (3%)
Query 48 LMQMIFDKAVSQHHFIQMYVMLCGKLKTDFQELLQDDHRS--TKFRRILIDQCEDSF 102
L +IFDKAV + F MY LC ++ ++ F+R+L++ C+ F
Sbjct 13 LTTLIFDKAVLEPTFCPMYAQLCFDIRHKMPRFPPSAPKTDEISFKRVLLNTCQKVF 69
> pfa:PF11_0465 dyn1; dynamin-like protein; K01528 dynamin GTPase
[EC:3.6.5.5]
Length=837
Score = 34.7 bits (78), Expect = 0.079, Method: Compositional matrix adjust.
Identities = 40/136 (29%), Positives = 59/136 (43%), Gaps = 26/136 (19%)
Query 10 VLLRHLKGLLNKLTIE-----RF--DRIYQQILNAGIKQEKEVKALMQMI---------- 52
VLLRH+K L + IE R+ D++Y+ N + K+ + L MI
Sbjct 291 VLLRHIKNFLPDIKIEINDKIRYINDKLYELGTNVPLDATKKTQLLWSMITDYCEIFKNT 350
Query 53 ----FDKAVSQHHFIQMYVMLCG-KLKTDFQELLQDDHRSTKFRRILIDQCEDSFNANLE 107
+DK V FI+ +LCG K++T F E L D++ L D D E
Sbjct 351 LKGKYDKRV--QVFIENNDILCGLKVRTIFNEFL-DEYVGKNVTSELTDNDIDDAICLHE 407
Query 108 PIAVPQGLSEDDAYEF 123
++P G D +EF
Sbjct 408 GDSLP-GFPSPDTFEF 422
> xla:380223 paip1, MGC154295, MGC53704; poly(A) binding protein
interacting protein 1; K14322 polyadenylate-binding protein-interacting
protein 1
Length=463
Score = 33.1 bits (74), Expect = 0.25, Method: Composition-based stats.
Identities = 16/72 (22%), Positives = 39/72 (54%), Gaps = 4/72 (5%)
Query 31 YQQILNAGIKQEKEVKALMQMIFDKAVSQHHFIQMYVMLCGKLKTDFQELLQDDHRSTKF 90
+ +LN + ++ ++ L+++I+ +A+S +F LC L + L + ++ F
Sbjct 169 FSDVLNNCVTTDESLQELVELIYQQAISVPNFSYTGARLCNYLSNN----LHINPQNQNF 224
Query 91 RRILIDQCEDSF 102
R++L+ +C+ F
Sbjct 225 RQLLLKRCQTEF 236
> cel:M110.4 ifg-1; Initiation Factor 4G (eIF4G) family member
(ifg-1); K03260 translation initiation factor 4G
Length=1156
Score = 32.0 bits (71), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 20/93 (21%), Positives = 51/93 (54%), Gaps = 4/93 (4%)
Query 11 LLRHLKGLLNKLTIERFDRIYQQILNAGIKQ-EKEVKALMQMIFDKAVSQHHFIQMYVML 69
+ + ++ L+NK+T + ++ ++ + + ++ +++++FDKAV + F +Y +
Sbjct 517 VCKKVRSLMNKVTPTSQRPLTEEFISYNVSSNDAQLAQVVEIVFDKAVEEPKFCALYAEM 576
Query 70 CGKLKTDFQELLQDDHRSTKFRRILIDQCEDSF 102
C K + + EL Q +S FR ++ + + +F
Sbjct 577 C-KAQAN-HELSQTGGKSA-FRNKVLTRTQMTF 606
> cel:C26C6.3 nas-36; Nematode AStacin protease family member
(nas-36); K08076 astacin [EC:3.4.24.21]
Length=617
Score = 30.8 bits (68), Expect = 1.0, Method: Composition-based stats.
Identities = 18/42 (42%), Positives = 23/42 (54%), Gaps = 1/42 (2%)
Query 62 FIQMYVMLCGKLKTDFQEL-LQDDHRSTKFRRILIDQCEDSF 102
FI+ + LC DF EL + DD R+T FR D+ E SF
Sbjct 413 FIEDFSFLCTSTCVDFVELKISDDLRNTGFRFCCYDKPEISF 454
> mmu:66053 Ppil2, 0610009L05Rik, 1700016N17Rik, 4921520K19Rik,
4930511F14Rik, AA589416, C130078A06Rik; peptidylprolyl isomerase
(cyclophilin)-like 2 (EC:5.2.1.8); K10598 peptidyl-prolyl
cis-trans isomerase-like 2 [EC:5.2.1.8]
Length=521
Score = 30.8 bits (68), Expect = 1.0, Method: Composition-based stats.
Identities = 19/60 (31%), Positives = 28/60 (46%), Gaps = 2/60 (3%)
Query 59 QHHFIQMYVMLCGKLKTDFQELLQDDHRSTKFRRILIDQCEDSFNANLEPIAVPQGLSED 118
QH +MY+ C + T F + D T FRR+ D C S + P+ P+G+ D
Sbjct 5 QHQKDKMYIT-CAEY-THFYGGRKPDISQTSFRRLPFDHCSLSLQPFVYPVCTPEGVVFD 62
> hsa:23759 PPIL2, CYC4, CYP60, Cyp-60, FLJ39930, MGC33174, MGC787,
hCyP-60; peptidylprolyl isomerase (cyclophilin)-like 2
(EC:5.2.1.8); K10598 peptidyl-prolyl cis-trans isomerase-like
2 [EC:5.2.1.8]
Length=520
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 19/60 (31%), Positives = 27/60 (45%), Gaps = 2/60 (3%)
Query 59 QHHFIQMYVMLCGKLKTDFQELLQDDHRSTKFRRILIDQCEDSFNANLEPIAVPQGLSED 118
QH +MY+ C + T F + D T FRR+ D C S + P+ P G+ D
Sbjct 5 QHQKDKMYIT-CAEY-THFYGGKKPDLPQTNFRRLPFDHCSLSLQPFVYPVCTPDGIVFD 62
> mmu:219148 Fam167a, A030013D21; family with sequence similarity
167, member A
Length=215
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 18/66 (27%), Positives = 34/66 (51%), Gaps = 0/66 (0%)
Query 12 LRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQHHFIQMYVMLCG 71
L L+G +NKL IE+ R+++++LN + +E L + D ++ + M + L G
Sbjct 142 LMRLRGDINKLKIEQTCRLHRRMLNDAAFELEERDELSDLFCDSPLASSFSLSMPLKLIG 201
Query 72 KLKTDF 77
K +
Sbjct 202 VTKMNI 207
> mmu:74763 Nat15, 1200013P24Rik, AI315146; N-acetyltransferase
15 (GCN5-related, putative)
Length=242
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 16/55 (29%), Positives = 25/55 (45%), Gaps = 0/55 (0%)
Query 59 QHHFIQMYVMLCGKLKTDFQELLQDDHRSTKFRRILIDQCEDSFNANLEPIAVPQ 113
QHH++ Y + G LK F +L + + + Q S ANL P ++P
Sbjct 157 QHHYLPYYYSIRGVLKDGFTYVLYINGGHPPWTILDYIQHLGSALANLSPCSIPH 211
> sce:YGR110W CLD1; Cld1p; K13535 cardiolipin-specific phospholipase
[EC:3.1.1.-]
Length=445
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 21/80 (26%), Positives = 40/80 (50%), Gaps = 3/80 (3%)
Query 11 LLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQ---MIFDKAVSQHHFIQMYV 67
L+R+ + L +K+T R ++ ILN +Q K +AL + IF+K S + + +
Sbjct 295 LVRNFRQLGSKITSGWSYRRFKHILNGDPEQSKRFEALHRYAYAIFNKRGSGEYLLSFAL 354
Query 68 MLCGKLKTDFQELLQDDHRS 87
G+ + ++ L D +S
Sbjct 355 KCGGEPRLSLEQQLFDGKKS 374
> pfa:MAL7P1.12 erythrocyte membrane-associated antigen
Length=2283
Score = 28.5 bits (62), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 18/70 (25%), Positives = 36/70 (51%), Gaps = 11/70 (15%)
Query 42 EKEVKALMQMIFDKAVSQHHFIQMYVMLCGKLKTDFQELLQDDHRSTKFRRILIDQCEDS 101
+KE K + ++F +AV +++ + + G E+ ++ KF R++ID+C S
Sbjct 1882 QKEAKVMKALLFLEAVKKYNVVIATCVGSG------HEIFDNE----KFERVIIDECAQS 1931
Query 102 FN-ANLEPIA 110
+NL P+
Sbjct 1932 IEPSNLIPLG 1941
> hsa:83648 FAM167A, C8orf13, D8S265, DKFZp761G151, MGC120649,
MGC120650, MGC120651; family with sequence similarity 167,
member A
Length=214
Score = 27.7 bits (60), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 17/66 (25%), Positives = 32/66 (48%), Gaps = 0/66 (0%)
Query 12 LRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQHHFIQMYVMLCG 71
L L+G +NKL IE R+++++LN + +E L + D ++ + + L G
Sbjct 141 LMRLRGDINKLKIEHTCRLHRRMLNDATYELEERDELADLFCDSPLASSFSLSTPLKLIG 200
Query 72 KLKTDF 77
K +
Sbjct 201 VTKMNI 206
> mmu:110596 Rgnef, 9230110L08Rik, AI323540, D13Bwg1089e, RIP2,
RhoGEF, Rhoip2, p190RhoGEF; Rho-guanine nucleotide exchange
factor
Length=1700
Score = 27.7 bits (60), Expect = 10.0, Method: Composition-based stats.
Identities = 14/48 (29%), Positives = 26/48 (54%), Gaps = 0/48 (0%)
Query 40 KQEKEVKALMQMIFDKAVSQHHFIQMYVMLCGKLKTDFQELLQDDHRS 87
+QEK+V +IF+ ++ H IQ +++ + +E LQ DH +
Sbjct 839 RQEKDVIKRQDVIFELMQTEVHHIQTLLIMSEVFRKGMKEELQLDHST 886
Lambda K H
0.322 0.137 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2099897216
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40