bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1081_orf1
Length=336
Score E
Sequences producing significant alignments: (Bits) Value
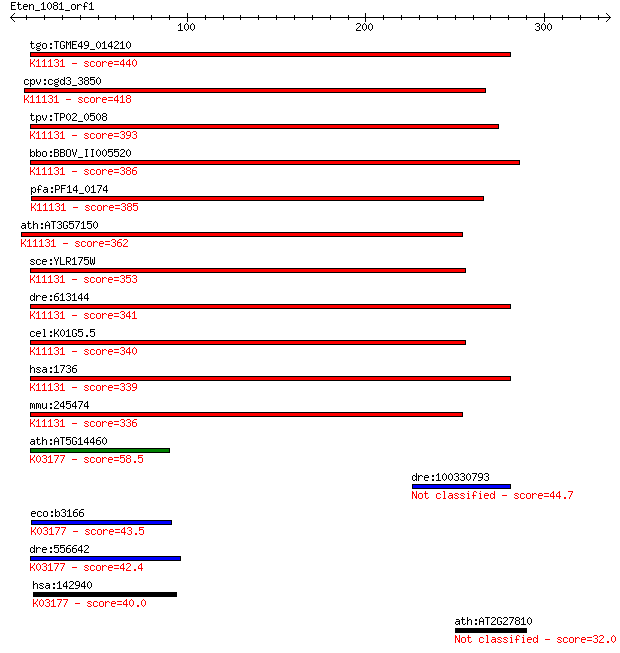
tgo:TGME49_014210 centromere/microtubule binding protein, puta... 440 4e-123
cpv:cgd3_3850 Cbf5p; centromere-binding factor 5 like PUA doma... 418 1e-116
tpv:TP02_0508 rRNA pseudouridine synthase; K11131 H/ACA ribonu... 393 5e-109
bbo:BBOV_II005520 18.m06459; rRNA pseudouridine synthase famil... 386 5e-107
pfa:PF14_0174 pseudouridine synthase, putative; K11131 H/ACA r... 385 8e-107
ath:AT3G57150 NAP57; NAP57 (Arabidopsis thaliana homologue of ... 362 1e-99
sce:YLR175W CBF5; Cbf5p (EC:5.4.99.-); K11131 H/ACA ribonucleo... 353 7e-97
dre:613144 dkc1, MGC110395, fv62a07, wu:fa28f10, wu:fc87a02, w... 341 2e-93
cel:K01G5.5 hypothetical protein; K11131 H/ACA ribonucleoprote... 340 6e-93
hsa:1736 DKC1, CBF5, DKC, FLJ97620, NAP57, NOLA4, XAP101; dysk... 339 1e-92
mmu:245474 Dkc1, BC068171, MGC107501; dyskeratosis congenita 1... 336 6e-92
ath:AT5G14460 pseudouridine synthase/ transporter; K03177 tRNA... 58.5 3e-08
dre:100330793 Dkc1 protein-like 44.7 5e-04
eco:b3166 truB, ECK3155, JW3135, yhbA; tRNA U55 pseudouridine ... 43.5 0.001
dre:556642 trub1, zgc:195155; TruB pseudouridine (psi) synthas... 42.4 0.002
hsa:142940 TRUB1, PUS4; TruB pseudouridine (psi) synthase homo... 40.0 0.013
ath:AT2G27810 xanthine/uracil permease family protein 32.0 3.7
> tgo:TGME49_014210 centromere/microtubule binding protein, putative
; K11131 H/ACA ribonucleoprotein complex subunit 4 [EC:5.4.99.-]
Length=505
Score = 440 bits (1132), Expect = 4e-123, Method: Compositional matrix adjust.
Identities = 211/273 (77%), Positives = 234/273 (85%), Gaps = 4/273 (1%)
Query 12 QALETLTGALFQRPPVISAVKRQLRIRTIYESEILSYDEDKHLCVFRVSCEAGTYIRTLC 71
+ALETLTGALFQRPPV++AVKRQLRIRTIY S +L YDE +H+ VF CEAGTYIRTLC
Sbjct 163 RALETLTGALFQRPPVVAAVKRQLRIRTIYASRLLDYDEQRHMAVFWAKCEAGTYIRTLC 222
Query 72 VHLGLLLGTGAHMQELRRVRSGNLQEDGNLVTLHDVLDAMFVYDAFRDETYLRRIIFPLE 131
VHLGL+LG GAHMQELRRVRSGNL E +LVT+HDVLDAM VYD+ RDETYLRR IFPLE
Sbjct 223 VHLGLVLGCGAHMQELRRVRSGNLGERDHLVTMHDVLDAMHVYDSTRDETYLRRTIFPLE 282
Query 132 MLLVGLPRIVVKDSCVNAICYGAKLMIPGVLRFENNIEVNAEVVLMTTKGEAIALGIAQM 191
+LL G PRIVVKDSCVNAICYGAKLMIPGVLRFE+ IEV+ EVVLMTTKGEAIALG+AQM
Sbjct 283 LLLTGFPRIVVKDSCVNAICYGAKLMIPGVLRFESGIEVHQEVVLMTTKGEAIALGVAQM 342
Query 192 STPVIASVDHGIVAAVKRVLMDRDTYAMRWGYGPRASEKKKMILGGLLDKKGRPNENTPK 251
++ VIA+VDHG+V+ VKRVLMDRDTY MRWGYGPRASEKKKMIL GLLDKKG+PNE TP
Sbjct 343 TSSVIATVDHGVVSTVKRVLMDRDTYDMRWGYGPRASEKKKMILAGLLDKKGKPNEKTPA 402
Query 252 EWLKTEGFLPKLTGEPEA----PAENAVKSERE 280
WLK+EG+LPKL G E E AVK E +
Sbjct 403 SWLKSEGYLPKLCGSEETENGEATEAAVKEEEQ 435
> cpv:cgd3_3850 Cbf5p; centromere-binding factor 5 like PUA domain
containing protein with a type I pseudouridine synthase
domain ; K11131 H/ACA ribonucleoprotein complex subunit 4 [EC:5.4.99.-]
Length=520
Score = 418 bits (1075), Expect = 1e-116, Method: Compositional matrix adjust.
Identities = 192/258 (74%), Positives = 228/258 (88%), Gaps = 0/258 (0%)
Query 9 ASMQALETLTGALFQRPPVISAVKRQLRIRTIYESEILSYDEDKHLCVFRVSCEAGTYIR 68
A + LETLTGALFQRPPVISAVKRQLR+R+IY+S++L YDE ++L VF CEAGTYIR
Sbjct 151 AVSRCLETLTGALFQRPPVISAVKRQLRVRSIYDSKLLDYDEKRNLAVFWAKCEAGTYIR 210
Query 69 TLCVHLGLLLGTGAHMQELRRVRSGNLQEDGNLVTLHDVLDAMFVYDAFRDETYLRRIIF 128
T+CVH+GLLLG GAHMQELRRVRSG L E+ N+VT+HD+LDA ++ D +RDE+YLRRII
Sbjct 211 TMCVHMGLLLGVGAHMQELRRVRSGCLSENDNMVTMHDILDAQYMLDNYRDESYLRRIIS 270
Query 129 PLEMLLVGLPRIVVKDSCVNAICYGAKLMIPGVLRFENNIEVNAEVVLMTTKGEAIALGI 188
PLE+LL LPRIVVKDSCVNAICYGAKLMIPGVLRF+N I+V EVV+MTTKGEAIA+G+
Sbjct 271 PLELLLTDLPRIVVKDSCVNAICYGAKLMIPGVLRFDNGIDVGTEVVMMTTKGEAIAIGV 330
Query 189 AQMSTPVIASVDHGIVAAVKRVLMDRDTYAMRWGYGPRASEKKKMILGGLLDKKGRPNEN 248
AQM+T VIASVDHG+VA +KRV+M+RDTY MRWG+GPRASEKKK+ILGG LDK G+PNE
Sbjct 331 AQMTTAVIASVDHGVVAIIKRVIMERDTYDMRWGHGPRASEKKKLILGGKLDKHGKPNEQ 390
Query 249 TPKEWLKTEGFLPKLTGE 266
TP+ WLK EG++PKLTG+
Sbjct 391 TPESWLKYEGYIPKLTGD 408
> tpv:TP02_0508 rRNA pseudouridine synthase; K11131 H/ACA ribonucleoprotein
complex subunit 4 [EC:5.4.99.-]
Length=421
Score = 393 bits (1010), Expect = 5e-109, Method: Compositional matrix adjust.
Identities = 179/262 (68%), Positives = 220/262 (83%), Gaps = 0/262 (0%)
Query 12 QALETLTGALFQRPPVISAVKRQLRIRTIYESEILSYDEDKHLCVFRVSCEAGTYIRTLC 71
+ L LTGALFQRPP+ISAVKRQLR+RTIY S++L Y+E + L +F V CEAGTY+RTLC
Sbjct 142 KVLAELTGALFQRPPLISAVKRQLRVRTIYASKLLDYNEQEDLALFWVKCEAGTYVRTLC 201
Query 72 VHLGLLLGTGAHMQELRRVRSGNLQEDGNLVTLHDVLDAMFVYDAFRDETYLRRIIFPLE 131
+H+GLLLGTGAHMQELRR++SGN+ E N+VT+HDVLD+ ++YD DE+YLR ++ PLE
Sbjct 202 IHMGLLLGTGAHMQELRRIKSGNISEYDNMVTMHDVLDSQYLYDTTGDESYLRYVVTPLE 261
Query 132 MLLVGLPRIVVKDSCVNAICYGAKLMIPGVLRFENNIEVNAEVVLMTTKGEAIALGIAQM 191
LL G R+VVKDSCVNAICYGAKLMIPGVLRFE NIE++ E+V++TTKGEAIALGIAQM
Sbjct 262 QLLTGHKRVVVKDSCVNAICYGAKLMIPGVLRFETNIELHDEIVMITTKGEAIALGIAQM 321
Query 192 STPVIASVDHGIVAAVKRVLMDRDTYAMRWGYGPRASEKKKMILGGLLDKKGRPNENTPK 251
++ VIA+VDHGIVA +KRV+MDRDTY +RWGYG R++EKK++I GLLDK G+ NENTP+
Sbjct 322 TSAVIATVDHGIVAVIKRVIMDRDTYPLRWGYGNRSAEKKRLISLGLLDKDGKVNENTPQ 381
Query 252 EWLKTEGFLPKLTGEPEAPAEN 273
W EG+LPKLTG P EN
Sbjct 382 SWTNNEGYLPKLTGTPTNNHEN 403
> bbo:BBOV_II005520 18.m06459; rRNA pseudouridine synthase family
protein; K11131 H/ACA ribonucleoprotein complex subunit
4 [EC:5.4.99.-]
Length=418
Score = 386 bits (992), Expect = 5e-107, Method: Compositional matrix adjust.
Identities = 180/274 (65%), Positives = 223/274 (81%), Gaps = 7/274 (2%)
Query 12 QALETLTGALFQRPPVISAVKRQLRIRTIYESEILSYDEDKHLCVFRVSCEAGTYIRTLC 71
+AL+TL+G LFQRPP+ISAVKRQLR+RTI+E++IL YDE + + F V CEAGTYIRTLC
Sbjct 142 KALQTLSGCLFQRPPLISAVKRQLRVRTIHETKILDYDESRCMATFWVRCEAGTYIRTLC 201
Query 72 VHLGLLLGTGAHMQELRRVRSGNLQEDGNLVTLHDVLDAMFVYDAFRDETYLRRIIFPLE 131
+HLGLLLG GAHMQELRRV+SG++ E N+VT+HDVLDA ++ D DE+Y+RR+I PLE
Sbjct 202 IHLGLLLGCGAHMQELRRVKSGHISEYDNMVTMHDVLDAQYLLDNTNDESYIRRVISPLE 261
Query 132 MLLVGLPRIVVKDSCVNAICYGAKLMIPGVLRFENNIEVNAEVVLMTTKGEAIALGIAQM 191
LL RIVVKDSCVNAICYGAKLM+PGVLRFEN IE++ ++VL+TTKGEAIALGIAQM
Sbjct 262 RLLTDYKRIVVKDSCVNAICYGAKLMVPGVLRFENGIELHEQIVLITTKGEAIALGIAQM 321
Query 192 STPVIASVDHGIVAAVKRVLMDRDTYAMRWGYGPRASEKKKMILGGLLDKKGRPNENTPK 251
T VIASVDHG+V+ +KRV+MDRDTY++RWG+GPRA+EKK++ GLLD+ G+PNE TP
Sbjct 322 PTAVIASVDHGVVSVIKRVIMDRDTYSLRWGFGPRATEKKRLQSAGLLDEHGKPNEKTPA 381
Query 252 EWLKTEGFLPKLTGEPEAPAENAVKSEREEGAPE 285
W K+EG+LP + GE K +EG PE
Sbjct 382 SWTKSEGYLPPMIGE-------DAKKRVQEGEPE 408
> pfa:PF14_0174 pseudouridine synthase, putative; K11131 H/ACA
ribonucleoprotein complex subunit 4 [EC:5.4.99.-]
Length=466
Score = 385 bits (990), Expect = 8e-107, Method: Compositional matrix adjust.
Identities = 171/253 (67%), Positives = 211/253 (83%), Gaps = 0/253 (0%)
Query 13 ALETLTGALFQRPPVISAVKRQLRIRTIYESEILSYDEDKHLCVFRVSCEAGTYIRTLCV 72
L GA+FQRPP+I AVKRQLR+RTIY+S++L YDE ++CVF V C+AGTYIRTLC
Sbjct 144 VLNNFQGAIFQRPPLICAVKRQLRVRTIYDSKLLDYDETNNVCVFWVKCQAGTYIRTLCE 203
Query 73 HLGLLLGTGAHMQELRRVRSGNLQEDGNLVTLHDVLDAMFVYDAFRDETYLRRIIFPLEM 132
H+GLLLG GAHMQELRRV+SGN+ E N+ TLHD+LDA ++YD DE+YLR+II PLE
Sbjct 204 HIGLLLGVGAHMQELRRVKSGNMTEYDNMCTLHDILDAQYIYDTTGDESYLRKIITPLEK 263
Query 133 LLVGLPRIVVKDSCVNAICYGAKLMIPGVLRFENNIEVNAEVVLMTTKGEAIALGIAQMS 192
LL+ PRIV+KDS VNAICYGAKL IPGVLRF+NNI+VN E+V+MTTKGEA+AL IAQM+
Sbjct 264 LLINFPRIVIKDSAVNAICYGAKLTIPGVLRFDNNIDVNTEIVIMTTKGEAVALAIAQMT 323
Query 193 TPVIASVDHGIVAAVKRVLMDRDTYAMRWGYGPRASEKKKMILGGLLDKKGRPNENTPKE 252
+ VIA+VDHG+V+ KRV+MDRDTY ++WG+G R+ EKKK+IL GLLDK G+PNE TP
Sbjct 324 STVIATVDHGVVSLTKRVIMDRDTYDVKWGFGNRSMEKKKLILAGLLDKYGKPNEKTPLS 383
Query 253 WLKTEGFLPKLTG 265
W+K+EG+ PKL G
Sbjct 384 WIKSEGYAPKLIG 396
> ath:AT3G57150 NAP57; NAP57 (Arabidopsis thaliana homologue of
NAP57); pseudouridine synthase; K11131 H/ACA ribonucleoprotein
complex subunit 4 [EC:5.4.99.-]
Length=565
Score = 362 bits (929), Expect = 1e-99, Method: Compositional matrix adjust.
Identities = 182/248 (73%), Positives = 213/248 (85%), Gaps = 1/248 (0%)
Query 7 DVASM-QALETLTGALFQRPPVISAVKRQLRIRTIYESEILSYDEDKHLCVFRVSCEAGT 65
DVA + +ALE+LTGA+FQRPP+ISAVKRQLRIRTIYES++L YD D+HL VF VSCEAGT
Sbjct 156 DVAKVARALESLTGAVFQRPPLISAVKRQLRIRTIYESKLLEYDADRHLVVFWVSCEAGT 215
Query 66 YIRTLCVHLGLLLGTGAHMQELRRVRSGNLQEDGNLVTLHDVLDAMFVYDAFRDETYLRR 125
YIRT+CVHLGLLLG G HMQELRRVRSG L E+ N+VT+HDV+DA FVYD RDE+YLRR
Sbjct 216 YIRTMCVHLGLLLGVGGHMQELRRVRSGILGENNNMVTMHDVMDAQFVYDNSRDESYLRR 275
Query 126 IIFPLEMLLVGLPRIVVKDSCVNAICYGAKLMIPGVLRFENNIEVNAEVVLMTTKGEAIA 185
+I PLEM+L R+VVKDS VNAICYGAKLMIPG+LRFEN+I+V EVVLMTTKGEAIA
Sbjct 276 VIMPLEMILTSYKRLVVKDSAVNAICYGAKLMIPGLLRFENDIDVGTEVVLMTTKGEAIA 335
Query 186 LGIAQMSTPVIASVDHGIVAAVKRVLMDRDTYAMRWGYGPRASEKKKMILGGLLDKKGRP 245
+GIA+M+T V+A+ DHG+VA +KRV+MDRDTY +WG GPRAS KKK+I G LDK G+P
Sbjct 336 VGIAEMTTSVMATCDHGVVAKIKRVVMDRDTYPRKWGLGPRASMKKKLIADGKLDKHGKP 395
Query 246 NENTPKEW 253
NE TP EW
Sbjct 396 NEKTPVEW 403
> sce:YLR175W CBF5; Cbf5p (EC:5.4.99.-); K11131 H/ACA ribonucleoprotein
complex subunit 4 [EC:5.4.99.-]
Length=483
Score = 353 bits (905), Expect = 7e-97, Method: Compositional matrix adjust.
Identities = 168/244 (68%), Positives = 200/244 (81%), Gaps = 0/244 (0%)
Query 12 QALETLTGALFQRPPVISAVKRQLRIRTIYESEILSYDEDKHLCVFRVSCEAGTYIRTLC 71
++LE LTGALFQRPP+ISAVKRQLR+RTIYES ++ +D ++L VF SCEAGTY+RTLC
Sbjct 141 RSLENLTGALFQRPPLISAVKRQLRVRTIYESNLIEFDNKRNLGVFWASCEAGTYMRTLC 200
Query 72 VHLGLLLGTGAHMQELRRVRSGNLQEDGNLVTLHDVLDAMFVYDAFRDETYLRRIIFPLE 131
VHLG+LLG G HMQELRRVRSG L E+ N+VTLHDV+DA +VYD RDE+YLR II PLE
Sbjct 201 VHLGMLLGVGGHMQELRRVRSGALSENDNMVTLHDVMDAQWVYDNTRDESYLRSIIQPLE 260
Query 132 MLLVGLPRIVVKDSCVNAICYGAKLMIPGVLRFENNIEVNAEVVLMTTKGEAIALGIAQM 191
LLVG RIVVKDS VNA+CYGAKLMIPG+LR+E IE+ E+VL+TTKGEAIA+ IAQM
Sbjct 261 TLLVGYKRIVVKDSAVNAVCYGAKLMIPGLLRYEEGIELYDEIVLITTKGEAIAVAIAQM 320
Query 192 STPVIASVDHGIVAAVKRVLMDRDTYAMRWGYGPRASEKKKMILGGLLDKKGRPNENTPK 251
ST +AS DHG+VA+VKR +M+RD Y RWG GP A +KK+M G LDK GR NENTP+
Sbjct 321 STVDLASCDHGVVASVKRCIMERDLYPRRWGLGPVAQKKKQMKADGKLDKYGRVNENTPE 380
Query 252 EWLK 255
+W K
Sbjct 381 QWKK 384
> dre:613144 dkc1, MGC110395, fv62a07, wu:fa28f10, wu:fc87a02,
wu:fi24a05, wu:fv62a07, zgc:110395; dyskeratosis congenita
1, dyskerin; K11131 H/ACA ribonucleoprotein complex subunit
4 [EC:5.4.99.-]
Length=506
Score = 341 bits (875), Expect = 2e-93, Method: Compositional matrix adjust.
Identities = 169/269 (62%), Positives = 215/269 (79%), Gaps = 2/269 (0%)
Query 12 QALETLTGALFQRPPVISAVKRQLRIRTIYESEILSYDEDKHLCVFRVSCEAGTYIRTLC 71
+ALE LTGALFQRPP+I+AVKRQLR+RTIYES+++ YD ++ L +F VSCEAGTYIRTLC
Sbjct 166 RALECLTGALFQRPPLIAAVKRQLRVRTIYESKLIEYDPERRLGIFWVSCEAGTYIRTLC 225
Query 72 VHLGLLLGTGAHMQELRRVRSGNLQEDGNLVTLHDVLDAMFVYDAFRDETYLRRIIFPLE 131
VHLGLLLG G MQELRRVRSG L E NLVT+HDVLDA + +D +DETYLRR+IFPLE
Sbjct 226 VHLGLLLGVGGQMQELRRVRSGVLGEKDNLVTMHDVLDAQWQFDHNKDETYLRRVIFPLE 285
Query 132 MLLVGLPRIVVKDSCVNAICYGAKLMIPGVLRFENNIEVNAEVVLMTTKGEAIALGIAQM 191
LL+ RIV+KDS VNAICYGAK+M+PGVLR+E+ IEVN ++V++TTKGEAI +A M
Sbjct 286 KLLISHKRIVMKDSAVNAICYGAKIMLPGVLRYEDGIEVNQDIVVITTKGEAICTAVALM 345
Query 192 STPVIASVDHGIVAAVKRVLMDRDTYAMRWGYGPRASEKKKMILGGLLDKKGRPNENTPK 251
+T VI++ DHG+VA +KRV+M+RDTY +WG GP+AS+KK MI GLLDK G+PN +TP
Sbjct 346 TTAVISTCDHGVVAKIKRVIMERDTYPRKWGLGPKASQKKMMIQKGLLDKHGKPNNSTPS 405
Query 252 EWLKTEGFLPKLTGEPEAPAENAVKSERE 280
+W EG++ T + + E + K +R+
Sbjct 406 DW--KEGYVDYSTTKVKKGGEASAKRKRD 432
> cel:K01G5.5 hypothetical protein; K11131 H/ACA ribonucleoprotein
complex subunit 4 [EC:5.4.99.-]
Length=445
Score = 340 bits (871), Expect = 6e-93, Method: Compositional matrix adjust.
Identities = 152/244 (62%), Positives = 196/244 (80%), Gaps = 0/244 (0%)
Query 12 QALETLTGALFQRPPVISAVKRQLRIRTIYESEILSYDEDKHLCVFRVSCEAGTYIRTLC 71
QALE LTGALFQRPP+ISAVKRQLRIRT+YE++ + YD + + +F CE+GTY+RT+C
Sbjct 159 QALEKLTGALFQRPPLISAVKRQLRIRTVYENKFIEYDPAQQMGIFNCICESGTYVRTIC 218
Query 72 VHLGLLLGTGAHMQELRRVRSGNLQEDGNLVTLHDVLDAMFVYDAFRDETYLRRIIFPLE 131
VHLGL+LG G MQELRR RSG E+ N+VT+HDVLDA ++ D +DE+Y+R I+ PLE
Sbjct 219 VHLGLILGCGGQMQELRRNRSGICDENENMVTMHDVLDAQYLLDTQKDESYMRHIVRPLE 278
Query 132 MLLVGLPRIVVKDSCVNAICYGAKLMIPGVLRFENNIEVNAEVVLMTTKGEAIALGIAQM 191
LL R+VVKDSC+NAICYGAK++IPG+LR++++IEV E+V+M+TKGEAI + IAQM
Sbjct 279 ALLTQHKRVVVKDSCINAICYGAKILIPGILRYDDDIEVGKEIVIMSTKGEAICIAIAQM 338
Query 192 STPVIASVDHGIVAAVKRVLMDRDTYAMRWGYGPRASEKKKMILGGLLDKKGRPNENTPK 251
+T IASVDHG+VA KRV+M+RD Y +WG GP AS+KK+M+ GLLDK G+PN+ TPK
Sbjct 339 NTSTIASVDHGVVAKSKRVIMERDVYGRKWGLGPVASKKKQMVKDGLLDKFGKPNDTTPK 398
Query 252 EWLK 255
W K
Sbjct 399 SWAK 402
> hsa:1736 DKC1, CBF5, DKC, FLJ97620, NAP57, NOLA4, XAP101; dyskeratosis
congenita 1, dyskerin; K11131 H/ACA ribonucleoprotein
complex subunit 4 [EC:5.4.99.-]
Length=509
Score = 339 bits (869), Expect = 1e-92, Method: Compositional matrix adjust.
Identities = 167/273 (61%), Positives = 217/273 (79%), Gaps = 4/273 (1%)
Query 12 QALETLTGALFQRPPVISAVKRQLRIRTIYESEILSYDEDKHLCVFRVSCEAGTYIRTLC 71
+ALETLTGALFQRPP+I+AVKRQLR+RTIYES+++ YD ++ L +F VSCEAGTYIRTLC
Sbjct 171 RALETLTGALFQRPPLIAAVKRQLRVRTIYESKMIEYDPERRLGIFWVSCEAGTYIRTLC 230
Query 72 VHLGLLLGTGAHMQELRRVRSGNLQEDGNLVTLHDVLDAMFVYDAFRDETYLRRIIFPLE 131
VHLGLLLG G MQELRRVRSG + E ++VT+HDVLDA ++YD +DE+YLRR+++PLE
Sbjct 231 VHLGLLLGVGGQMQELRRVRSGVMSEKDHMVTMHDVLDAQWLYDNHKDESYLRRVVYPLE 290
Query 132 MLLVGLPRIVVKDSCVNAICYGAKLMIPGVLRFENNIEVNAEVVLMTTKGEAIALGIAQM 191
LL R+V+KDS VNAICYGAK+M+PGVLR+E+ IEVN E+V++TTKGEAI + IA M
Sbjct 291 KLLTSHKRLVMKDSAVNAICYGAKIMLPGVLRYEDGIEVNQEIVVITTKGEAICMAIALM 350
Query 192 STPVIASVDHGIVAAVKRVLMDRDTYAMRWGYGPRASEKKKMILGGLLDKKGRPNENTPK 251
+T VI++ DHGIVA +KRV+M+RDTY +WG GP+AS+KK MI GLLDK G+P ++TP
Sbjct 351 TTAVISTCDHGIVAKIKRVIMERDTYPRKWGLGPKASQKKLMIKQGLLDKHGKPTDSTPA 410
Query 252 EWLKTEGFLPKLTGE----PEAPAENAVKSERE 280
W + E ++ E P+ AE A ++R+
Sbjct 411 TWKQDESAKKEVVAEVVKAPQVVAEAAKTAKRK 443
> mmu:245474 Dkc1, BC068171, MGC107501; dyskeratosis congenita
1, dyskerin homolog (human); K11131 H/ACA ribonucleoprotein
complex subunit 4 [EC:5.4.99.-]
Length=509
Score = 336 bits (862), Expect = 6e-92, Method: Compositional matrix adjust.
Identities = 160/242 (66%), Positives = 203/242 (83%), Gaps = 0/242 (0%)
Query 12 QALETLTGALFQRPPVISAVKRQLRIRTIYESEILSYDEDKHLCVFRVSCEAGTYIRTLC 71
+ALETLTGALFQRPP+I+AVKRQLR+RTIYES+++ YD ++ L +F VSCEAGTYIRTLC
Sbjct 171 RALETLTGALFQRPPLIAAVKRQLRVRTIYESKMIEYDPERRLGIFWVSCEAGTYIRTLC 230
Query 72 VHLGLLLGTGAHMQELRRVRSGNLQEDGNLVTLHDVLDAMFVYDAFRDETYLRRIIFPLE 131
VHLGLLLG G MQELRRVRSG + E ++VT+HDVLDA ++YD +DE+YLRR+++PLE
Sbjct 231 VHLGLLLGVGGQMQELRRVRSGVMSEKDHMVTMHDVLDAQWLYDNHKDESYLRRVVYPLE 290
Query 132 MLLVGLPRIVVKDSCVNAICYGAKLMIPGVLRFENNIEVNAEVVLMTTKGEAIALGIAQM 191
LL R+V+KDS VNAICYGAK+M+PG+LR+E+ IEVN E+V++TTKGEAI + IA M
Sbjct 291 KLLTSHKRLVMKDSAVNAICYGAKIMLPGLLRYEDGIEVNQEIVVITTKGEAICMAIALM 350
Query 192 STPVIASVDHGIVAAVKRVLMDRDTYAMRWGYGPRASEKKKMILGGLLDKKGRPNENTPK 251
+T VI++ DHGIVA +KRV+M+RDTY +WG GP+AS+KK MI GLLDK G+P +NTP
Sbjct 351 TTAVISTCDHGIVAKIKRVIMERDTYPRKWGLGPKASQKKMMIKQGLLDKHGKPTDNTPA 410
Query 252 EW 253
W
Sbjct 411 TW 412
> ath:AT5G14460 pseudouridine synthase/ transporter; K03177 tRNA
pseudouridine synthase B [EC:5.4.99.12]
Length=540
Score = 58.5 bits (140), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 33/95 (34%), Positives = 50/95 (52%), Gaps = 17/95 (17%)
Query 12 QALETLTGALFQRPPVISAVKR---------------QLRIR--TIYESEILSYDEDKHL 54
+AL + G ++Q PP+ SA+K +L R +I++ +I +D+
Sbjct 420 KALTSFLGEIWQVPPMFSAIKVGGEKMYEKARRGETVELSPRRISIFQFDIERSLDDRQN 479
Query 55 CVFRVSCEAGTYIRTLCVHLGLLLGTGAHMQELRR 89
+FRV C GTYIR+LC L LG+ AH+ LRR
Sbjct 480 LIFRVICSKGTYIRSLCADLAKALGSCAHLTALRR 514
> dre:100330793 Dkc1 protein-like
Length=138
Score = 44.7 bits (104), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 23/55 (41%), Positives = 35/55 (63%), Gaps = 2/55 (3%)
Query 226 RASEKKKMILGGLLDKKGRPNENTPKEWLKTEGFLPKLTGEPEAPAENAVKSERE 280
+AS+KK MI GLLDK G+PN +TP +W EG++ T + + E + K +R+
Sbjct 12 QASQKKMMIQKGLLDKHGKPNNSTPSDW--KEGYVDYSTTKVKKGGEASAKRKRD 64
> eco:b3166 truB, ECK3155, JW3135, yhbA; tRNA U55 pseudouridine
synthase (EC:4.2.1.70); K03177 tRNA pseudouridine synthase
B [EC:5.4.99.12]
Length=314
Score = 43.5 bits (101), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 31/95 (32%), Positives = 46/95 (48%), Gaps = 19/95 (20%)
Query 13 ALETLTGALFQRPPVISAVK-----------------RQLRIRTIYESEILSYDEDKHLC 55
AL+T G + Q P + SA+K R+ R T+YE + ++ ++
Sbjct 111 ALDTFRGDIEQIPSMYSALKYQGKKLYEYARQGIEVPREARPITVYELLFIRHEGNE--L 168
Query 56 VFRVSCEAGTYIRTLCVHLGLLLGTGAHMQELRRV 90
+ C GTYIRT+ LG LG GAH+ LRR+
Sbjct 169 ELEIHCSKGTYIRTIIDDLGEKLGCGAHVIYLRRL 203
> dre:556642 trub1, zgc:195155; TruB pseudouridine (psi) synthase
homolog 1 (E. coli); K03177 tRNA pseudouridine synthase
B [EC:5.4.99.12]
Length=290
Score = 42.4 bits (98), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 43/102 (42%), Gaps = 20/102 (19%)
Query 12 QALETLTGALFQRPPVISAVKRQ------------------LRIRTIYESEILSYDEDKH 53
+ ++ TG + Q PP+ SA+K+ R T+Y + +
Sbjct 133 EKMKQFTGEIMQVPPLYSALKKDGKRMSVLLKQGQEVEAKPARPVTVYNLSLEDFSPP-- 190
Query 54 LCVFRVSCEAGTYIRTLCVHLGLLLGTGAHMQELRRVRSGNL 95
L V C G Y+R+L LG L + AH++EL R + G
Sbjct 191 LFTIDVECGGGFYVRSLIDDLGKALSSCAHIKELTRTKQGQF 232
> hsa:142940 TRUB1, PUS4; TruB pseudouridine (psi) synthase homolog
1 (E. coli); K03177 tRNA pseudouridine synthase B [EC:5.4.99.12]
Length=349
Score = 40.0 bits (92), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 27/96 (28%), Positives = 47/96 (48%), Gaps = 16/96 (16%)
Query 14 LETLTGALFQRPPVISAVKRQ-LRIRTIYE-SEILSYDEDKHLCVFRVS----------- 60
L+ TG + Q PP+ SA+K+ R+ T+ + E++ + + V+ +S
Sbjct 186 LQKFTGNIMQVPPLYSALKKDGQRLSTLMKRGEVVEAKPARPVTVYSISLQKFQPPFFTL 245
Query 61 ---CEAGTYIRTLCVHLGLLLGTGAHMQELRRVRSG 93
C G YIR+L +G L + A++ EL R + G
Sbjct 246 DVECGGGFYIRSLVSDIGKELSSCANVLELTRTKQG 281
> ath:AT2G27810 xanthine/uracil permease family protein
Length=542
Score = 32.0 bits (71), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 24/46 (52%), Gaps = 6/46 (13%)
Query 250 PKEWLKTEGFLPKLTGEPEAPAENA------VKSEREEGAPECEEG 289
P W K GF PK +GE A ++ V+++++E P+ E G
Sbjct 26 PSSWAKKTGFRPKFSGETTATDSSSGQLSLPVRAKQQETQPDLEAG 71
Lambda K H
0.317 0.135 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 14156784820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40