bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1061_orf1
Length=106
Score E
Sequences producing significant alignments: (Bits) Value
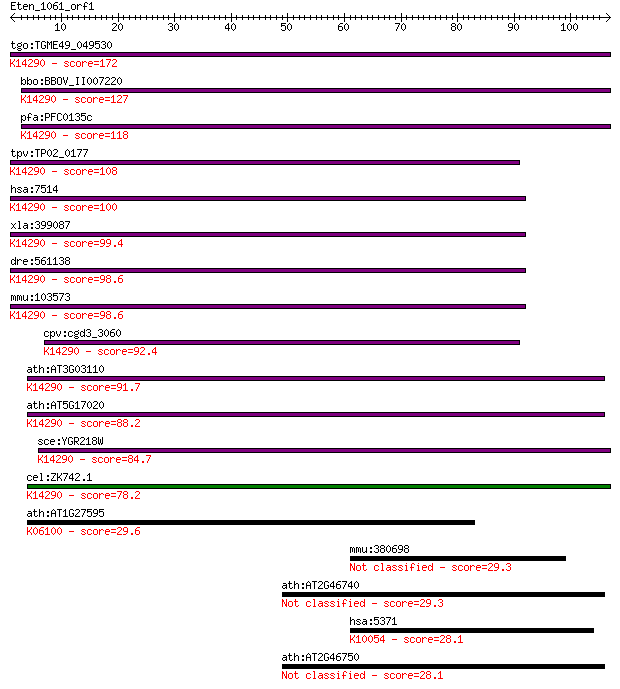
tgo:TGME49_049530 exportin, putative ; K14290 exportin-1 172 2e-43
bbo:BBOV_II007220 18.m06600; exportin 1; K14290 exportin-1 127 7e-30
pfa:PFC0135c exportin 1, putative; K14290 exportin-1 118 5e-27
tpv:TP02_0177 importin beta-related nuclear transport factor; ... 108 4e-24
hsa:7514 XPO1, CRM1, DKFZp686B1823, emb; exportin 1 (CRM1 homo... 100 1e-21
xla:399087 xpo1, crm1, exportin-1; exportin 1 (CRM1 homolog); ... 99.4 2e-21
dre:561138 xpo1b, xpo1; exportin 1 (CRM1 homolog, yeast) b; K1... 98.6 4e-21
mmu:103573 Xpo1, AA420417, Crm1, Exp1; exportin 1, CRM1 homolo... 98.6 4e-21
cpv:cgd3_3060 exportin 1 ; K14290 exportin-1 92.4 3e-19
ath:AT3G03110 XPO1B; XPO1B; binding / protein transporter; K14... 91.7 5e-19
ath:AT5G17020 XPO1A; XPO1A; protein binding / protein transpor... 88.2 6e-18
sce:YGR218W CRM1, KAP124, XPO1; Crm1p; K14290 exportin-1 84.7 6e-17
cel:ZK742.1 xpo-1; eXPOrtin (nuclear export receptor) family m... 78.2 6e-15
ath:AT1G27595 hypothetical protein; K06100 symplekin 29.6 2.2
mmu:380698 Obscn, BC046431, Gm878, MGC51514, OTTMUSG0000000578... 29.3 2.8
ath:AT2G46740 FAD-binding domain-containing protein 29.3 3.3
hsa:5371 PML, MYL, PP8675, RNF71, TRIM19; promyelocytic leukem... 28.1 6.6
ath:AT2G46750 FAD-binding domain-containing protein 28.1 8.0
> tgo:TGME49_049530 exportin, putative ; K14290 exportin-1
Length=1125
Score = 172 bits (437), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 78/106 (73%), Positives = 94/106 (88%), Gaps = 0/106 (0%)
Query 1 TSVLPVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFV 60
+ VLPVIF+++F+ TL MIK+DFQSYPDHRE+FY LLKA NQHCF GLF+LP+ QLKA+V
Sbjct 868 SPVLPVIFEFVFESTLNMIKTDFQSYPDHREKFYELLKACNQHCFDGLFALPAHQLKAYV 927
Query 61 ESLVWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCNYYF 106
ESLVWAFKHEHPS+AE+GLQVT+EFL +LI KREVL+DFC +YF
Sbjct 928 ESLVWAFKHEHPSVAEQGLQVTYEFLLKLINDKREVLSDFCNLFYF 973
> bbo:BBOV_II007220 18.m06600; exportin 1; K14290 exportin-1
Length=1186
Score = 127 bits (320), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 62/116 (53%), Positives = 78/116 (67%), Gaps = 13/116 (11%)
Query 3 VLPVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVES 62
VLP IF+ IFDCTL M+K DF S+P+HRE FY +L+ +HCF GL LPS +L+A+V S
Sbjct 905 VLPQIFEQIFDCTLDMVKMDFHSFPEHREYFYEMLQKCTKHCFDGLLLLPSERLRAYVMS 964
Query 63 LVWAFKHEHPSLAEEGLQVTHEFLQRLI------------EGKREVLNDFCCNYYF 106
L+WAFKHEHPS+AE GL V EFL L+ +G VL+ FC NYY+
Sbjct 965 LIWAFKHEHPSVAERGLTVVREFLNNLMLLDRRNDQSPGGQGMSAVLS-FCRNYYY 1019
> pfa:PFC0135c exportin 1, putative; K14290 exportin-1
Length=1254
Score = 118 bits (295), Expect = 5e-27, Method: Compositional matrix adjust.
Identities = 50/104 (48%), Positives = 73/104 (70%), Gaps = 0/104 (0%)
Query 3 VLPVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVES 62
+LP + +Y+ T+ MIK+DF SYP+HRE+FY L A +HCF LF+L S F++S
Sbjct 995 ILPTVLNYVLLPTIDMIKNDFSSYPEHREKFYNFLDACVRHCFDYLFTLDSEIFNTFIQS 1054
Query 63 LVWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCNYYF 106
L+WA KHEHPS+A+ GL++T +FL +I K+E L +FC +Y+
Sbjct 1055 LLWAIKHEHPSVADHGLRITQQFLHNIIIKKKEYLEEFCKAFYY 1098
> tpv:TP02_0177 importin beta-related nuclear transport factor;
K14290 exportin-1
Length=1067
Score = 108 bits (270), Expect = 4e-24, Method: Composition-based stats.
Identities = 48/90 (53%), Positives = 66/90 (73%), Gaps = 0/90 (0%)
Query 1 TSVLPVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFV 60
+SVL IF+Y+FD +L M+K DF +YPDHRE FY +L A + F + LP+ +L+ FV
Sbjct 971 SSVLVQIFNYVFDTSLDMVKMDFHAYPDHREYFYEMLHKATKCSFDSVLLLPNERLRDFV 1030
Query 61 ESLVWAFKHEHPSLAEEGLQVTHEFLQRLI 90
SLVWAFKHEHPS+A+ GL +T EF++ +I
Sbjct 1031 MSLVWAFKHEHPSIADRGLLITLEFMKNII 1060
> hsa:7514 XPO1, CRM1, DKFZp686B1823, emb; exportin 1 (CRM1 homolog,
yeast); K14290 exportin-1
Length=1071
Score = 100 bits (248), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 41/91 (45%), Positives = 61/91 (67%), Gaps = 0/91 (0%)
Query 1 TSVLPVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFV 60
T+ +P IFD +F+CTL MI DF+ YP+HR F+ LL+A N HCF ++P TQ K +
Sbjct 816 TAEIPQIFDAVFECTLNMINKDFEEYPEHRTNFFLLLQAVNSHCFPAFLAIPPTQFKLVL 875
Query 61 ESLVWAFKHEHPSLAEEGLQVTHEFLQRLIE 91
+S++WAFKH ++A+ GLQ+ LQ + +
Sbjct 876 DSIIWAFKHTMRNVADTGLQILFTLLQNVAQ 906
> xla:399087 xpo1, crm1, exportin-1; exportin 1 (CRM1 homolog);
K14290 exportin-1
Length=1071
Score = 99.4 bits (246), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 40/91 (43%), Positives = 61/91 (67%), Gaps = 0/91 (0%)
Query 1 TSVLPVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFV 60
T+ +P IFD +F+CTL MI DF+ YP+HR F+ LL+A N HCF ++P Q K +
Sbjct 816 TAEIPQIFDAVFECTLNMINKDFEEYPEHRTHFFLLLQAVNSHCFPAFLAIPPAQFKLVL 875
Query 61 ESLVWAFKHEHPSLAEEGLQVTHEFLQRLIE 91
+S++WAFKH ++A+ GLQ+ + LQ + +
Sbjct 876 DSIIWAFKHTMRNVADTGLQILYTLLQNVAQ 906
> dre:561138 xpo1b, xpo1; exportin 1 (CRM1 homolog, yeast) b;
K14290 exportin-1
Length=1071
Score = 98.6 bits (244), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 40/91 (43%), Positives = 60/91 (65%), Gaps = 0/91 (0%)
Query 1 TSVLPVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFV 60
T +P IFD +F+CTL MI DF+ YP+HR F+ LL+A N HCF ++P Q K +
Sbjct 816 TGEIPQIFDAVFECTLNMINKDFEEYPEHRTHFFYLLQAVNSHCFPAFLAIPPAQFKLVL 875
Query 61 ESLVWAFKHEHPSLAEEGLQVTHEFLQRLIE 91
+S++WAFKH ++A+ GLQ+ + LQ + +
Sbjct 876 DSIIWAFKHTMRNVADTGLQILYTLLQNVAQ 906
> mmu:103573 Xpo1, AA420417, Crm1, Exp1; exportin 1, CRM1 homolog
(yeast); K14290 exportin-1
Length=1071
Score = 98.6 bits (244), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 40/91 (43%), Positives = 60/91 (65%), Gaps = 0/91 (0%)
Query 1 TSVLPVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFV 60
T+ +P IFD +F+CTL MI DF+ YP+HR F+ LL+A N HCF ++P Q K +
Sbjct 816 TAEIPQIFDAVFECTLNMINKDFEEYPEHRTNFFLLLQAVNSHCFPAFLAIPPAQFKLVL 875
Query 61 ESLVWAFKHEHPSLAEEGLQVTHEFLQRLIE 91
+S++WAFKH ++A+ GLQ+ LQ + +
Sbjct 876 DSIIWAFKHTMRNVADTGLQILFTLLQNVAQ 906
> cpv:cgd3_3060 exportin 1 ; K14290 exportin-1
Length=1266
Score = 92.4 bits (228), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 41/84 (48%), Positives = 58/84 (69%), Gaps = 0/84 (0%)
Query 7 IFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESLVWA 66
I ++F+ TL MIK +F +YPDHRE FY+ L N+ CF LF+LP L ++ES++WA
Sbjct 969 IIYHLFESTLSMIKDNFHAYPDHREFFYSFLADCNEFCFLQLFNLPGNILTLYIESIIWA 1028
Query 67 FKHEHPSLAEEGLQVTHEFLQRLI 90
+HE P++AE+GL V + FL LI
Sbjct 1029 IRHEQPNMAEKGLIVLYNFLINLI 1052
> ath:AT3G03110 XPO1B; XPO1B; binding / protein transporter; K14290
exportin-1
Length=1076
Score = 91.7 bits (226), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 41/102 (40%), Positives = 65/102 (63%), Gaps = 6/102 (5%)
Query 4 LPVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESL 63
+P+IF+ +F CTL+MI +F+ YP+HR +F++LL+A CF L L S QLK ++S+
Sbjct 823 VPLIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRAIATFCFRALIQLSSEQLKLVMDSV 882
Query 64 VWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCNYY 105
+WAF+H ++AE GL + E L+ + +DFC +Y
Sbjct 883 IWAFRHTERNIAETGLNLLLEMLKNFQK------SDFCNKFY 918
> ath:AT5G17020 XPO1A; XPO1A; protein binding / protein transporter/
receptor; K14290 exportin-1
Length=1075
Score = 88.2 bits (217), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 40/102 (39%), Positives = 64/102 (62%), Gaps = 6/102 (5%)
Query 4 LPVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESL 63
+P IF+ +F CTL+MI +F+ YP+HR +F++LL+A CF L L S QLK ++S+
Sbjct 822 VPHIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRAIATFCFPALIKLSSPQLKLVMDSI 881
Query 64 VWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCNYY 105
+WAF+H ++AE GL + E L+ + ++FC +Y
Sbjct 882 IWAFRHTERNIAETGLNLLLEMLKNFQQ------SEFCNQFY 917
> sce:YGR218W CRM1, KAP124, XPO1; Crm1p; K14290 exportin-1
Length=1084
Score = 84.7 bits (208), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 39/102 (38%), Positives = 57/102 (55%), Gaps = 1/102 (0%)
Query 6 VIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESLVW 65
+I +F+CTL MI DF YP+HR FY LLK N+ F+ LP K FV+++ W
Sbjct 832 LILQSVFECTLDMINKDFTEYPEHRVEFYKLLKVINEKSFAAFLELPPAAFKLFVDAICW 891
Query 66 AFKHEHPSLAEEGLQVTHEFLQRLIE-GKREVLNDFCCNYYF 106
AFKH + + GLQ+ + ++ + G N+F NY+F
Sbjct 892 AFKHNNRDVEVNGLQIALDLVKNIERMGNVPFANEFHKNYFF 933
> cel:ZK742.1 xpo-1; eXPOrtin (nuclear export receptor) family
member (xpo-1); K14290 exportin-1
Length=1080
Score = 78.2 bits (191), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 33/103 (32%), Positives = 56/103 (54%), Gaps = 0/103 (0%)
Query 4 LPVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESL 63
+P I +F C++ MI D +++P+HR F+ L+ + Q CF +P L ++++
Sbjct 829 VPSILSAVFQCSIDMINKDMEAFPEHRTNFFELVLSLVQECFPVFMEMPPEDLGTVIDAV 888
Query 64 VWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCNYYF 106
VWAF+H ++AE GL + E L R+ E ++ F YY
Sbjct 889 VWAFQHTMRNVAEIGLDILKELLARVSEQDDKIAQPFYKRYYI 931
> ath:AT1G27595 hypothetical protein; K06100 symplekin
Length=961
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 19/79 (24%), Positives = 30/79 (37%), Gaps = 4/79 (5%)
Query 4 LPVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESL 63
P + D++ + ++++ P F + H F L LP QL ES+
Sbjct 847 FPTLVDFVMEILSKLVRKQIWRLPKLWPGFLKCVSQTKPHSFPVLLELPVPQL----ESI 902
Query 64 VWAFKHEHPSLAEEGLQVT 82
+ F PSL Q T
Sbjct 903 MKKFPDLRPSLTAYANQPT 921
> mmu:380698 Obscn, BC046431, Gm878, MGC51514, OTTMUSG00000005786,
UNC89; obscurin, cytoskeletal calmodulin and titin-interacting
RhoGEF (EC:2.7.11.1)
Length=8025
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 18/38 (47%), Positives = 22/38 (57%), Gaps = 3/38 (7%)
Query 61 ESLVWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLN 98
ESLV H H SLAE G T EFLQ+L E+++
Sbjct 5736 ESLVAEEAHSHLSLAEVG---TEEFLQKLTSQITEMVS 5770
> ath:AT2G46740 FAD-binding domain-containing protein
Length=590
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 20/63 (31%), Positives = 30/63 (47%), Gaps = 8/63 (12%)
Query 49 FSLPSTQLKAFVESLVWAFKHEHPSLAEEGLQVTHEFLQRLIE------GKREVLNDFCC 102
FS+P TQ+K+F+ + K + SL GL++ + L R + GK DF
Sbjct 403 FSIPLTQVKSFINDIKSLLKIDSKSLC--GLELYYGILMRYVTSSPAYLGKETEAIDFDI 460
Query 103 NYY 105
YY
Sbjct 461 TYY 463
> hsa:5371 PML, MYL, PP8675, RNF71, TRIM19; promyelocytic leukemia;
K10054 probable transcription factor PML
Length=633
Score = 28.1 bits (61), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 21/43 (48%), Gaps = 4/43 (9%)
Query 61 ESLVWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCN 103
E+ W KHE LAE Q EFL +G R+ N FC N
Sbjct 153 EAHQWFLKHEARPLAELRNQSVREFL----DGTRKTNNIFCSN 191
> ath:AT2G46750 FAD-binding domain-containing protein
Length=591
Score = 28.1 bits (61), Expect = 8.0, Method: Composition-based stats.
Identities = 20/63 (31%), Positives = 29/63 (46%), Gaps = 8/63 (12%)
Query 49 FSLPSTQLKAFVESLVWAFKHEHPSLAEEGLQVTHEFLQRLIE------GKREVLNDFCC 102
S+P TQ+K+F+ + K E SL GL++ + L R + GK DF
Sbjct 399 LSVPLTQVKSFISDIKSLVKIEQKSLC--GLELHYGILMRYVTSSPAYLGKETEALDFDI 456
Query 103 NYY 105
YY
Sbjct 457 TYY 459
Lambda K H
0.328 0.140 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2012750684
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40