bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
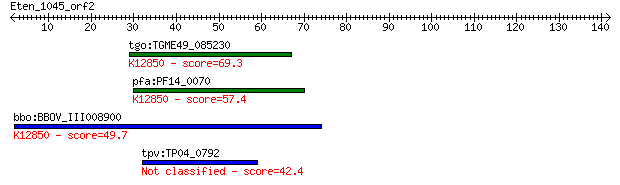
Query= Eten_1045_orf2
Length=141
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_085230 PRP38 family domain-containing protein ; K12... 69.3 4e-12
pfa:PF14_0070 pre-mRNA splicing factor, putative; K12850 pre-m... 57.4 1e-08
bbo:BBOV_III008900 17.m07776; PRP38 family protein; K12850 pre... 49.7 3e-06
tpv:TP04_0792 hypothetical protein 42.4 5e-04
> tgo:TGME49_085230 PRP38 family domain-containing protein ; K12850
pre-mRNA-splicing factor 38B
Length=614
Score = 69.3 bits (168), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 33/38 (86%), Positives = 36/38 (94%), Gaps = 0/38 (0%)
Query 29 AKTQQQLLQEFRRKEQEKALATGKDYARRPTSYKSALS 66
AKTQ LL+EFRRKE+EKALATGKDYARRPTSYKS+LS
Sbjct 470 AKTQHDLLKEFRRKEREKALATGKDYARRPTSYKSSLS 507
> pfa:PF14_0070 pre-mRNA splicing factor, putative; K12850 pre-mRNA-splicing
factor 38B
Length=690
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 27/40 (67%), Positives = 35/40 (87%), Gaps = 0/40 (0%)
Query 30 KTQQQLLQEFRRKEQEKALATGKDYARRPTSYKSALSLKM 69
KT+ +L+ F++ E +KALATGKDYARRPTSYKS+LSLK+
Sbjct 576 KTEDELISRFKKLESQKALATGKDYARRPTSYKSSLSLKV 615
> bbo:BBOV_III008900 17.m07776; PRP38 family protein; K12850 pre-mRNA-splicing
factor 38B
Length=483
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 45/72 (62%), Gaps = 1/72 (1%)
Query 2 PKAKRSRNRDGDKERRRSRSRSAERSAAKTQQQLLQEFRRKEQEKALATGKDYARRPTSY 61
P KR R+ D R+ S+ R ++++ L +E++R+E ++ LA GKDY +RP+S+
Sbjct 411 PGNKRHRDESIDDRDRKGTGNSSVRHG-ESREALFREYKRRESDRVLAVGKDYGKRPSSF 469
Query 62 KSALSLKMTSSR 73
++L KM ++R
Sbjct 470 NASLPSKMHTNR 481
> tpv:TP04_0792 hypothetical protein
Length=211
Score = 42.4 bits (98), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 16/27 (59%), Positives = 24/27 (88%), Gaps = 0/27 (0%)
Query 32 QQQLLQEFRRKEQEKALATGKDYARRP 58
++ L++EF+R+E+EKALA GKDY +RP
Sbjct 169 KEDLIREFKRREREKALAVGKDYGKRP 195
Lambda K H
0.309 0.120 0.321
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2618291680
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40