bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1024_orf3
Length=266
Score E
Sequences producing significant alignments: (Bits) Value
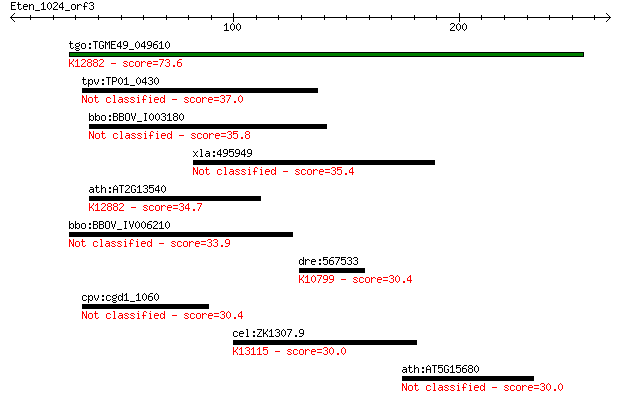
tgo:TGME49_049610 nuclear cap binding protein, putative ; K128... 73.6 7e-13
tpv:TP01_0430 hypothetical protein 37.0 0.074
bbo:BBOV_I003180 19.m02162; ABC transporter, ATP-binding prote... 35.8 0.15
xla:495949 ankrd16; ankyrin repeat domain 16 35.4
ath:AT2G13540 ABH1; ABH1 (ABA HYPERSENSITIVE 1); RNA cap bindi... 34.7 0.38
bbo:BBOV_IV006210 23.m06135; hypothetical protein 33.9 0.69
dre:567533 wu:fe02c12; si:ch211-155m12.3; K10799 tankyrase [EC... 30.4 6.3
cpv:cgd1_1060 hypothetical protein 30.4 7.1
cel:ZK1307.9 hypothetical protein; K13115 coiled-coil domain-c... 30.0 8.9
ath:AT5G15680 binding 30.0 9.1
> tgo:TGME49_049610 nuclear cap binding protein, putative ; K12882
nuclear cap-binding protein subunit 1
Length=1201
Score = 73.6 bits (179), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 67/287 (23%), Positives = 115/287 (40%), Gaps = 66/287 (22%)
Query 27 DTSGAPWTPEEALKLFVFCLLSFGSKTQTHLNRILSNYLQTFICFANQSEDPA------- 79
D G WT + ++ VF LL+ G KT TH R++ NY F + A
Sbjct 887 DEEGHLWTLTDLSQVLVFALLARGLKTLTHSERLVENYFPVFQFLKKGASHKAFLEMRPA 946
Query 80 -------------------------------EAREPEVDIHPAVLQAVQKFWSTSQQRTA 108
+A ++ A L+A+ FW S+Q+T
Sbjct 947 TLEAAGDDEDARDGDDSDTEMEEETDEEGLTKAERRSQEVEEAFLKAIFDFWKHSRQKTV 1006
Query 109 LTLHAFLKSGILQRARVLQ----ELCAADANIRDSWGQLELVETVLRGALDECENAREEA 164
LT+ F + ++ + V++ L D RD +EL + +L+ ++ E E+ +E A
Sbjct 1007 LTVRHFQRFDVVSKEGVVRFVFDSLTPTD---RDDSRVMELFDLLLQLSIQEFESKKETA 1063
Query 165 -----------------AAASAPMPTNTEAEQLLHFLMVHLIEDLIKETSPARSRFLFLR 207
A SA + + LLHF++V + +L++E AR L R
Sbjct 1064 LQLSQDCSDDAERNARGKAVSAAVEALEAVQTLLHFIVVGFMRELLREDQAARRVSLHAR 1123
Query 208 CIFFGRKYAEFINLQKLKEEVEMVLSGIEDRRVSNLLIVIGQVQKHY 254
++ R Y +I++ +L + ++ ++ LL V QVQ Y
Sbjct 1124 LLYLARDYGRYIDVSRLHVHIGDAMTD----ELNQLLTVSSQVQTQY 1166
> tpv:TP01_0430 hypothetical protein
Length=981
Score = 37.0 bits (84), Expect = 0.074, Method: Composition-based stats.
Identities = 31/107 (28%), Positives = 49/107 (45%), Gaps = 12/107 (11%)
Query 33 WTPEEALKLFVFCLLSFGSKTQTHLNRIL---SNYLQTFICFANQSEDPAEAREPEVDIH 89
W ++ + LF LL FGSK+ THL R+L S+ L+ F E P E P +
Sbjct 756 WKRDDLILLFWDTLLIFGSKSMTHLYRLLEFHSDVLKMF----ETPESPFEESLP----Y 807
Query 90 PAVLQAVQKFWSTSQQRTALTLHAFLKSGILQRARVLQELCAADANI 136
+ Q V F +R L+ F+++ + +L+ L AN+
Sbjct 808 KVMWQTVVTF-ERDHKRLELSFDFFIRNNVFTCPMILKFLFTLPANV 853
> bbo:BBOV_I003180 19.m02162; ABC transporter, ATP-binding protein
domain containing protein
Length=1555
Score = 35.8 bits (81), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 29/107 (27%), Positives = 47/107 (43%), Gaps = 14/107 (13%)
Query 36 EEALKLFVFCLLSFG-SKTQTHLNRILSNYLQTFICFANQSEDPAEAREPEVDIHPAVLQ 94
++A++LFV CL+ S++ + + +YL T A VD A ++
Sbjct 136 KDAMELFVVCLMRLVVSRSSLYSGSAILSYLGTAFILAGIVT---------VD---AFIE 183
Query 95 AVQKFWSTSQQ-RTALTLHAFLKSGILQRARVLQELCAADANIRDSW 140
A F+ R +TLH L IL R RV+ C +D + D +
Sbjct 184 AYHHFYYRRLSLRLEITLHTLLFGKILARDRVISSRCLSDGTVADEY 230
> xla:495949 ankrd16; ankyrin repeat domain 16
Length=293
Score = 35.4 bits (80), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 29/117 (24%), Positives = 44/117 (37%), Gaps = 10/117 (8%)
Query 82 REPEVDIHPAVLQAVQKFWSTSQQRTALTLHAFLKSGILQRARVLQELCAADANIRDSWG 141
RE +V I +L W T + LH G + VL E C D + +DS G
Sbjct 154 REGDVAIIQYLLDVFPDIWKTESKIKRTPLHTAAMHGCFEVIEVLLERCNYDPDCKDSCG 213
Query 142 QLELVETVLRGALD----------ECENAREEAAAASAPMPTNTEAEQLLHFLMVHL 188
++ V G L C +A + A + + T + L +L+ HL
Sbjct 214 VTPFMDAVQNGHLSIAQLLIEKKKVCYSAFDRMGAQALHLAAVTGQNESLQYLVSHL 270
> ath:AT2G13540 ABH1; ABH1 (ABA HYPERSENSITIVE 1); RNA cap binding;
K12882 nuclear cap-binding protein subunit 1
Length=848
Score = 34.7 bits (78), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 34/78 (43%), Gaps = 15/78 (19%)
Query 36 EEALKLFVFCLLSFGSKTQTHLNRILSNYLQTF--ICFANQSEDPAEAREPEVDIHPAVL 93
E L + V LL GSK+ THL +L Y Q F +C P+ D +L
Sbjct 532 EVTLTIVVQTLLDIGSKSFTHLVTVLERYGQVFSKLC-------------PDNDKQVMLL 578
Query 94 QAVQKFWSTSQQRTALTL 111
V +W + Q TA+ +
Sbjct 579 SQVSTYWKNNVQMTAVAI 596
> bbo:BBOV_IV006210 23.m06135; hypothetical protein
Length=991
Score = 33.9 bits (76), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 30/106 (28%), Positives = 50/106 (47%), Gaps = 14/106 (13%)
Query 27 DTSGAP----WTPEEALKLFVFCLLSFGSKTQTHLNRILSNY---LQTFICFANQSEDPA 79
D G P W+ +E + +F +L FG+K+ THL R++ + L+ FI SE+
Sbjct 753 DKDGQPTVKTWSRDELILIFWESVLLFGNKSLTHLLRLIEFHGEVLKNFI-----SEEQN 807
Query 80 EAREPEVDIHPAVLQAVQKFWSTSQQRTALTLHAFLKSGILQRARV 125
A E + VL ++ T ++ L + ++ GIL A V
Sbjct 808 GAFEDSISFKIMVL--TRETLKTDTKKFELVIDNLIREGILPPADV 851
> dre:567533 wu:fe02c12; si:ch211-155m12.3; K10799 tankyrase [EC:2.4.2.30]
Length=1267
Score = 30.4 bits (67), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 17/30 (56%), Gaps = 1/30 (3%)
Query 129 LC-AADANIRDSWGQLELVETVLRGALDEC 157
LC AD N RD+W L E ++G +D C
Sbjct 211 LCSGADPNARDNWNYTPLHEAAIKGKIDVC 240
> cpv:cgd1_1060 hypothetical protein
Length=1055
Score = 30.4 bits (67), Expect = 7.1, Method: Composition-based stats.
Identities = 13/57 (22%), Positives = 30/57 (52%), Gaps = 1/57 (1%)
Query 33 WTPEEALKLFVFCLLSFGSKTQTHLNRILSNYLQTFI-CFANQSEDPAEAREPEVDI 88
WT + L ++ GSK+ +H+ R+ +NY + + + ++ + E+ E + +I
Sbjct 710 WTKDSLFDLLFISIVDQGSKSPSHIRRLFANYKTSILEWYEDEKNNLTESSESDFNI 766
> cel:ZK1307.9 hypothetical protein; K13115 coiled-coil domain-containing
protein 130
Length=369
Score = 30.0 bits (66), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 36/81 (44%), Gaps = 6/81 (7%)
Query 100 WSTSQQRTALTLHAFLKSGILQRARVLQELCAADANIRDSWGQLELVETVLRGALDECEN 159
W + R T ++FL++ + L E A DAN+RD +L + T L L E E
Sbjct 171 WIQERMRDDFTANSFLRAQFRNEKKSLNETRARDANLRD---KLSIGTTQL---LPETEE 224
Query 160 AREEAAAASAPMPTNTEAEQL 180
R A+ + T T + L
Sbjct 225 DRRIASMMTRYRDTKTHDDHL 245
> ath:AT5G15680 binding
Length=2153
Score = 30.0 bits (66), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 34/74 (45%), Gaps = 18/74 (24%)
Query 175 TEAEQLLHFLMVHLIEDLIKETSPARSRFLFLRC----------------IFFGRKYAEF 218
T AE L F M H++ +L+K+ SP+ ++ + LR IF G +
Sbjct 392 TIAEHNLDFAMNHMLLELLKQDSPSEAKIIGLRALLALVMSPSSQYVGLEIFKGHGIGHY 451
Query 219 INLQKLKEEVEMVL 232
I K+K +E +L
Sbjct 452 I--PKVKAAIESIL 463
Lambda K H
0.320 0.133 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9890911460
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40