bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1022_orf1
Length=299
Score E
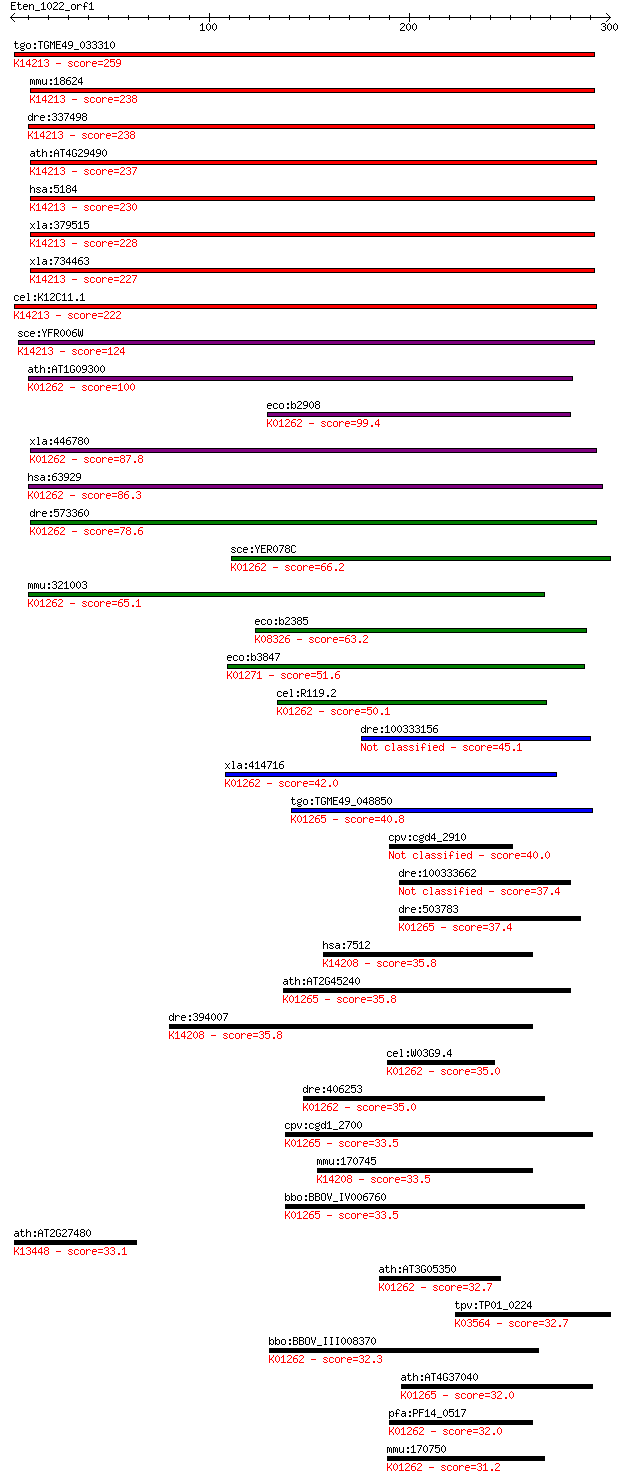
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_033310 prolidase, putative (EC:3.4.13.9); K14213 Xa... 259 7e-69
mmu:18624 Pepd, Pep-4, Pep4; peptidase D (EC:3.4.13.9); K14213... 238 1e-62
dre:337498 pepd, cb1000, fj78g11, wu:fj78g11; peptidase D (EC:... 238 2e-62
ath:AT4G29490 aminopeptidase/ manganese ion binding; K14213 Xa... 237 5e-62
hsa:5184 PEPD, MGC10905, PROLIDASE; peptidase D (EC:3.4.13.9);... 230 5e-60
xla:379515 hypothetical protein MGC64570; K14213 Xaa-Pro dipep... 228 2e-59
xla:734463 pepd, MGC115123; peptidase D (EC:3.4.13.9); K14213 ... 227 4e-59
cel:K12C11.1 hypothetical protein; K14213 Xaa-Pro dipeptidase ... 222 1e-57
sce:YFR006W Putative X-Pro aminopeptidase; green fluorescent p... 124 4e-28
ath:AT1G09300 metallopeptidase M24 family protein; K01262 Xaa-... 100 9e-21
eco:b2908 pepP, ECK2903, JW2876; proline aminopeptidase P II (... 99.4 1e-20
xla:446780 xpnpep3, MGC80423; X-prolyl aminopeptidase (aminope... 87.8 5e-17
hsa:63929 XPNPEP3, APP3, NPHPL1; X-prolyl aminopeptidase (amin... 86.3 1e-16
dre:573360 xpnpep3, MGC76905, zgc:76905; X-prolyl aminopeptida... 78.6 3e-14
sce:YER078C ICP55; Icp55p (EC:3.4.-.-); K01262 Xaa-Pro aminope... 66.2 1e-10
mmu:321003 Xpnpep3, APP3, E430012M05Rik; X-prolyl aminopeptida... 65.1 3e-10
eco:b2385 ypdF, ECK2381, JW2382; Xaa-Pro aminopeptidase; K0832... 63.2 1e-09
eco:b3847 pepQ, ECK3839, JW3823; proline dipeptidase (EC:3.4.1... 51.6 3e-06
cel:R119.2 hypothetical protein; K01262 Xaa-Pro aminopeptidase... 50.1 1e-05
dre:100333156 peptidase-like 45.1 3e-04
xla:414716 hypothetical protein MGC83093; K01262 Xaa-Pro amino... 42.0 0.003
tgo:TGME49_048850 methionine aminopeptidase, putative (EC:3.4.... 40.8 0.007
cpv:cgd4_2910 aminopeptidase 40.0 0.011
dre:100333662 methionyl aminopeptidase 1-like 37.4 0.065
dre:503783 metap1, im:7047238, wu:fc84e12, zgc:110093; methion... 37.4 0.070
hsa:7512 XPNPEP2, APP2; X-prolyl aminopeptidase (aminopeptidas... 35.8 0.18
ath:AT2G45240 MAP1A; MAP1A (METHIONINE AMINOPEPTIDASE 1A); ami... 35.8 0.22
dre:394007 xpnpep2, MGC63528, zgc:63528; X-prolyl aminopeptida... 35.8 0.22
cel:W03G9.4 app-1; AminoPeptidase P family member (app-1); K01... 35.0 0.31
dre:406253 xpnpep1, MGC56366, fa02e02, wu:fa02e02, zgc:56366, ... 35.0 0.34
cpv:cgd1_2700 methionine aminopeptidase with MYND finger at N-... 33.5 0.96
mmu:170745 Xpnpep2, 9030008G12Rik, mAPP; X-prolyl aminopeptida... 33.5 1.0
bbo:BBOV_IV006760 23.m06237; methionine aminopeptidase (EC:3.4... 33.5 1.0
ath:AT2G27480 calcium ion binding; K13448 calcium-binding prot... 33.1 1.4
ath:AT3G05350 aminopeptidase/ hydrolase; K01262 Xaa-Pro aminop... 32.7 1.7
tpv:TP01_0224 hypothetical protein; K03564 peroxiredoxin Q/BCP... 32.7 1.9
bbo:BBOV_III008370 17.m07732; metallopeptidase M24 family prot... 32.3 2.1
ath:AT4G37040 MAP1D; MAP1D (METHIONINE AMINOPEPTIDASE 1D); ami... 32.0 2.6
pfa:PF14_0517 peptidase, putative; K01262 Xaa-Pro aminopeptida... 32.0 2.6
mmu:170750 Xpnpep1, D230045I08Rik; X-prolyl aminopeptidase (am... 31.2 5.0
> tgo:TGME49_033310 prolidase, putative (EC:3.4.13.9); K14213
Xaa-Pro dipeptidase [EC:3.4.13.9]
Length=525
Score = 259 bits (663), Expect = 7e-69, Method: Compositional matrix adjust.
Identities = 135/289 (46%), Positives = 174/289 (60%), Gaps = 50/289 (17%)
Query 3 SADCNKAAFRQEAFFQHLVGVNEPDIYAAFVLNSHELLLFIPKIPADALRFMGPSKDADF 62
SADC+KA FRQE FF++L GVNE D++ + + +LF+P + RFMGP + A++
Sbjct 145 SADCDKAVFRQEQFFRYLFGVNEADVFGLLDFSRRQAVLFVPWTSPEYQRFMGPPRAAEW 204
Query 63 YRSRYAVTDAIAVEAISDVEAELQRRGVRTVHVLKGVNSDSGRPVAPPKALNSFSSFTVD 122
Y +QR G+ V K
Sbjct 205 Y---------------------MQRYGLDGAVVYK------------------------- 218
Query 123 CSALYDILVECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYVGF 182
L +I E KT LE +YL AACL SSQ H FV RNI GMVEGQ EA+F+A+V +
Sbjct 219 -EGLQEIREE---LKTELERDYLRAACLVSSQGHTFVMRNIYPGMVEGQGEALFRAFVHY 274
Query 183 VGGARHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTY 242
GGARHVAYDCICCAG + +ILHYGHAGRPNDG IK D+LLFDMGG+Y GY+TDIT +Y
Sbjct 275 AGGARHVAYDCICCAGPHGAILHYGHAGRPNDGVIKCGDMLLFDMGGEYGGYSTDITLSY 334
Query 243 PSNGIFTETQKAIYDAVLDAQKSVIERMRPGVEWTELHRLAELVILKHL 291
P NG+ + Q+ +Y+A +AQ++V M+PGV WT++HRLAE IL+ L
Sbjct 335 PVNGVCSREQRVVYEAAYEAQRAVEMAMKPGVMWTDMHRLAEKKILERL 383
> mmu:18624 Pepd, Pep-4, Pep4; peptidase D (EC:3.4.13.9); K14213
Xaa-Pro dipeptidase [EC:3.4.13.9]
Length=493
Score = 238 bits (608), Expect = 1e-62, Method: Compositional matrix adjust.
Identities = 120/281 (42%), Positives = 177/281 (62%), Gaps = 4/281 (1%)
Query 11 FRQEAFFQHLVGVNEPDIYAAFVLNSHELLLFIPKIPADALRFMGPSKDADFYRSRYAVT 70
FRQE+FF GV E Y +++ + LF+P++P +MG ++++ +YAV
Sbjct 65 FRQESFFHWAFGVVESGCYGVIDVDTGKSTLFVPRLPDSYATWMGKIHSKEYFKEKYAVD 124
Query 71 DAIAVEAISDVEAELQRRGVRTVHVLKGVNSDSGRPVAPPKALNSFSSFTVDCSALYDIL 130
D + I+ V L R + L+GVN+DSG V + S F V+ + L+ +
Sbjct 125 DVQYTDEIASV---LTSRNPSVLLTLRGVNTDSG-SVCREASFEGISKFNVNNTILHPEI 180
Query 131 VECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYVGFVGGARHVA 190
VECR+ KT +E E L SS+AH V + ++ GM E + E++F+ Y GG RH +
Sbjct 181 VECRVFKTDMELEVLRYTNRISSEAHREVMKAVKVGMKEYEMESLFQHYCYSRGGMRHTS 240
Query 191 YDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTE 250
Y CICC+GENA++LHYGHAG PND +IKD D+ LFDMGG+Y +A+DIT ++P+NG FTE
Sbjct 241 YTCICCSGENAAVLHYGHAGAPNDRTIKDGDICLFDMGGEYYCFASDITCSFPANGKFTE 300
Query 251 TQKAIYDAVLDAQKSVIERMRPGVEWTELHRLAELVILKHL 291
QKAIY+AVL + ++V+ M+PGV W ++HRLA+ + L+ L
Sbjct 301 DQKAIYEAVLRSCRTVMSTMKPGVWWPDMHRLADRIHLEEL 341
> dre:337498 pepd, cb1000, fj78g11, wu:fj78g11; peptidase D (EC:3.4.13.9);
K14213 Xaa-Pro dipeptidase [EC:3.4.13.9]
Length=496
Score = 238 bits (608), Expect = 2e-62, Method: Compositional matrix adjust.
Identities = 121/282 (42%), Positives = 179/282 (63%), Gaps = 4/282 (1%)
Query 10 AFRQEAFFQHLVGVNEPDIYAAFVLNSHELLLFIPKIPADALRFMGPSKDADFYRSRYAV 69
FRQE+FF GV E D Y A ++S + LLF+PK+P +MG ++ +YAV
Sbjct 65 TFRQESFFHWSFGVTEADCYGAIDVDSKKSLLFVPKLPESYATWMGEIFPPGHFKEKYAV 124
Query 70 TDAIAVEAISDVEAELQRRGVRTVHVLKGVNSDSGRPVAPPKALNSFSSFTVDCSALYDI 129
+ I+DV A+++ + T L+G+N+DSG + S F V+ S L+ +
Sbjct 125 DEVHFTTDIADVLAKMKPSVLLT---LRGLNTDSGS-TCREASFKGISRFEVNNSLLHPV 180
Query 130 LVECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYVGFVGGARHV 189
+VECRL KT +E E L SS+AH V R ++ G+ E + E++F+ Y GG RH
Sbjct 181 IVECRLLKTDMELEVLRYTNRISSEAHKEVMRRVKPGLKEYEMESLFQHYCYSRGGMRHT 240
Query 190 AYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFT 249
+Y CIC +G N+SILHYGHAG PND +I+D D+ LFDMGG+Y Y++DIT ++P+NG FT
Sbjct 241 SYTCICGSGNNSSILHYGHAGAPNDKTIQDGDMCLFDMGGEYYCYSSDITCSFPANGKFT 300
Query 250 ETQKAIYDAVLDAQKSVIERMRPGVEWTELHRLAELVILKHL 291
Q+ IY+AVL + ++V+ ++PGV+WT++HRLA+ V L+ L
Sbjct 301 ADQRTIYEAVLKSSRAVMAAIKPGVKWTDMHRLADRVHLEEL 342
> ath:AT4G29490 aminopeptidase/ manganese ion binding; K14213
Xaa-Pro dipeptidase [EC:3.4.13.9]
Length=486
Score = 237 bits (604), Expect = 5e-62, Method: Compositional matrix adjust.
Identities = 118/283 (41%), Positives = 170/283 (60%), Gaps = 2/283 (0%)
Query 11 FRQEAFFQHLVGVNEPDIYAAFVLNSHELLLFIPKIPADALRFMGPSKDADFYRSRYAVT 70
FRQE++F +L GV EPD Y A + S + +LFIP++P D ++G K ++ Y V
Sbjct 59 FRQESYFAYLFGVREPDFYGAIDIGSGKSILFIPRLPDDYAVWLGEIKPLSHFKETYMVD 118
Query 71 DAIAVEAISDV-EAELQRRGVRTVHVLKGVNSDSGRPVAPPKALNSFSSFTVDCSALYDI 129
V+ I V + + G +++L G+N+DS + P + F D + L+ I
Sbjct 119 MVFYVDEIIQVFNEQFKGSGKPLLYLLHGLNTDSSN-FSKPASFEGIDKFETDLTTLHPI 177
Query 130 LVECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYVGFVGGARHV 189
L ECR+ K+ LE + + A SS+AH V R + GM E Q E++F + GG RH
Sbjct 178 LAECRVIKSSLELQLIQFANDISSEAHIEVMRKVTPGMKEYQMESMFLHHSYMYGGCRHC 237
Query 190 AYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFT 249
+Y CIC G+N+++LHYGHA PND + +D DL L DMG +Y+ Y +DIT ++P NG FT
Sbjct 238 SYTCICATGDNSAVLHYGHAAAPNDRTFEDGDLALLDMGAEYHFYGSDITCSFPVNGKFT 297
Query 250 ETQKAIYDAVLDAQKSVIERMRPGVEWTELHRLAELVILKHLK 292
Q IY+AVLDA SVI M+PGV W ++H+LAE +IL+ LK
Sbjct 298 SDQSLIYNAVLDAHNSVISAMKPGVNWVDMHKLAEKIILESLK 340
> hsa:5184 PEPD, MGC10905, PROLIDASE; peptidase D (EC:3.4.13.9);
K14213 Xaa-Pro dipeptidase [EC:3.4.13.9]
Length=493
Score = 230 bits (586), Expect = 5e-60, Method: Compositional matrix adjust.
Identities = 116/281 (41%), Positives = 178/281 (63%), Gaps = 4/281 (1%)
Query 11 FRQEAFFQHLVGVNEPDIYAAFVLNSHELLLFIPKIPADALRFMGPSKDADFYRSRYAVT 70
FRQE+FF GV EP Y +++ + LF+P++PA +MG + ++ +YAV
Sbjct 65 FRQESFFHWAFGVTEPGCYGVIDVDTGKSTLFVPRLPASHATWMGKIHSKEHFKEKYAVD 124
Query 71 DAIAVEAISDVEAELQRRGVRTVHVLKGVNSDSGRPVAPPKALNSFSSFTVDCSALYDIL 130
D V+ I+ V L + + L+GVN+DSG V + + S F V+ + L+ +
Sbjct 125 DVQYVDEIASV---LTSQKPSVLLTLRGVNTDSGS-VCREASFDGISKFEVNNTILHPEI 180
Query 131 VECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYVGFVGGARHVA 190
VECR+ KT +E E L SS+AH V + ++ GM E + E++F+ Y GG RH +
Sbjct 181 VECRVFKTDMELEVLRYTNKISSEAHREVMKAVKVGMKEYELESLFEHYCYSRGGMRHSS 240
Query 191 YDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTE 250
Y CIC +GEN+++LHYGHAG PND +I++ D+ LFDMGG+Y +A+DIT ++P+NG FT
Sbjct 241 YTCICGSGENSAVLHYGHAGAPNDRTIQNGDMCLFDMGGEYYCFASDITCSFPANGKFTA 300
Query 251 TQKAIYDAVLDAQKSVIERMRPGVEWTELHRLAELVILKHL 291
QKA+Y+AVL + ++V+ M+PGV W ++HRLA+ + L+ L
Sbjct 301 DQKAVYEAVLRSSRAVMGAMKPGVWWPDMHRLADRIHLEEL 341
> xla:379515 hypothetical protein MGC64570; K14213 Xaa-Pro dipeptidase
[EC:3.4.13.9]
Length=498
Score = 228 bits (581), Expect = 2e-59, Method: Compositional matrix adjust.
Identities = 116/281 (41%), Positives = 176/281 (62%), Gaps = 4/281 (1%)
Query 11 FRQEAFFQHLVGVNEPDIYAAFVLNSHELLLFIPKIPADALRFMGPSKDADFYRSRYAVT 70
FRQE+FF GV E Y A +N+ + +LFIPK+P +MG + ++ +YA+
Sbjct 65 FRQESFFHWTFGVIEAGCYGAVDVNTGKSILFIPKLPESYAVWMGKIHPPEHFKEKYAID 124
Query 71 DAIAVEAISDVEAELQRRGVRTVHVLKGVNSDSGRPVAPPKALNSFSSFTVDCSALYDIL 130
+ + D+ + L+ + + L+GVN+DSG V + S F+V+ + L+ +
Sbjct 125 E---IYFTCDISSVLKAKTPSVLLTLRGVNTDSGS-VCKEASFEGISEFSVNNTLLHPEI 180
Query 131 VECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYVGFVGGARHVA 190
VECR+ KT +E E L SS+AH V + R GM E + E++F Y GG RH +
Sbjct 181 VECRVFKTDMELEVLRYTNRISSEAHKEVMKAARVGMKEFELESVFLHYCYARGGMRHTS 240
Query 191 YDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTE 250
Y CIC +G+N+SILHYGHAG PND ++ D D+ LFDMGG+Y Y++DIT ++P+NG FT
Sbjct 241 YTCICGSGDNSSILHYGHAGAPNDKTVTDGDMCLFDMGGEYYCYSSDITCSFPANGKFTP 300
Query 251 TQKAIYDAVLDAQKSVIERMRPGVEWTELHRLAELVILKHL 291
Q+A+Y+AVL + ++VI+ ++PGV W ++HRLA+ V L+ L
Sbjct 301 DQRAVYEAVLKSSRAVIKAVKPGVAWPDMHRLADRVHLEEL 341
> xla:734463 pepd, MGC115123; peptidase D (EC:3.4.13.9); K14213
Xaa-Pro dipeptidase [EC:3.4.13.9]
Length=498
Score = 227 bits (578), Expect = 4e-59, Method: Compositional matrix adjust.
Identities = 113/281 (40%), Positives = 176/281 (62%), Gaps = 4/281 (1%)
Query 11 FRQEAFFQHLVGVNEPDIYAAFVLNSHELLLFIPKIPADALRFMGPSKDADFYRSRYAVT 70
FRQE+FF GV EP Y A +++ + + FIPK+P +MG + ++ +YA+
Sbjct 65 FRQESFFHWTFGVTEPGCYGAVDVDTGKSIAFIPKLPESYAVWMGKIHPPEHFKEKYAID 124
Query 71 DAIAVEAISDVEAELQRRGVRTVHVLKGVNSDSGRPVAPPKALNSFSSFTVDCSALYDIL 130
+ + D+ + L+ + + L+GVN+DSG V + S F+V+ + L+ +
Sbjct 125 E---IYFTCDISSVLKAKTPSVLLTLRGVNTDSGS-VCKEASFEGISEFSVNNTLLHPEI 180
Query 131 VECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYVGFVGGARHVA 190
VECR+ KT +E E L SS+AH V + R GM E + E++F Y GG RH +
Sbjct 181 VECRVFKTDMELEVLQYTNRISSEAHKEVMKAARVGMKEYELESVFLHYCYARGGMRHTS 240
Query 191 YDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTE 250
Y CIC +G+N+S+LHYGHAG PND ++ D D+ LFDMGG+Y Y++DIT ++P+NG FT
Sbjct 241 YTCICGSGDNSSVLHYGHAGAPNDKTVMDGDMCLFDMGGEYYCYSSDITCSFPANGKFTP 300
Query 251 TQKAIYDAVLDAQKSVIERMRPGVEWTELHRLAELVILKHL 291
Q+A+Y+AVL + ++V++ ++PGV W ++HRLA+ V L+ L
Sbjct 301 DQRAVYEAVLKSSRAVMKAVKPGVAWPDMHRLADRVHLEEL 341
> cel:K12C11.1 hypothetical protein; K14213 Xaa-Pro dipeptidase
[EC:3.4.13.9]
Length=498
Score = 222 bits (565), Expect = 1e-57, Method: Compositional matrix adjust.
Identities = 116/291 (39%), Positives = 173/291 (59%), Gaps = 2/291 (0%)
Query 3 SADCNKAAFRQEAFFQHLVGVNEPDIYAAF-VLNSHELLLFIPKIPADALRFMGPSKDAD 61
+ D FRQE++F GVNE + Y A V + + LF P++ + G +
Sbjct 50 NTDAADLPFRQESYFFWTFGVNESEFYGAIDVRSGGKTTLFAPRLDPSYAIWDGKINNEQ 109
Query 62 FYRSRYAVTDAIAVEAISDVEAELQRRGVRTVHVLKGVNSDSGRPVAPPKALNSFSSFTV 121
F++ +YAV + + + + + +L+ + V++L+ N+DSG +A PK S F +
Sbjct 110 FFKEKYAVDEVVFNDKTTTIAEKLKELSAKHVYLLRAENTDSGDVLAEPKFAGS-GDFQL 168
Query 122 DCSALYDILVECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYVG 181
D LY + E R+ KT E + A +S+AH +++R G+ E Q E++F+
Sbjct 169 DTELLYKEMAELRVVKTEKEIGVMRYASKIASEAHRAAMKHMRPGLYEYQLESLFRHTSY 228
Query 182 FVGGARHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFT 241
+ GG RH+AY CI G N S+LHYGHA PND IKD D+ LFDMG +YN YA+DIT +
Sbjct 229 YHGGCRHLAYTCIAATGCNGSVLHYGHANAPNDKFIKDGDMCLFDMGPEYNCYASDITTS 288
Query 242 YPSNGIFTETQKAIYDAVLDAQKSVIERMRPGVEWTELHRLAELVILKHLK 292
+PSNG FTE QK +Y+AVL A +V++ +PGV WT++H L+E VIL+HLK
Sbjct 289 FPSNGKFTEKQKIVYNAVLAANLAVLKAAKPGVRWTDMHILSEKVILEHLK 339
> sce:YFR006W Putative X-Pro aminopeptidase; green fluorescent
protein (GFP)-fusion protein localizes to the cytoplasm; YFR006W
is not an essential gene (EC:3.4.-.-); K14213 Xaa-Pro
dipeptidase [EC:3.4.13.9]
Length=535
Score = 124 bits (312), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 85/287 (29%), Positives = 134/287 (46%), Gaps = 18/287 (6%)
Query 5 DCNKAAFRQEAFFQHLVGVNEPDIYAAFVLNSHELLLFIPKIPADALRFMGPSKDADFYR 64
D NK FRQ +F HL GV+ P F ++ +L LF+P I + + + G D
Sbjct 113 DTNKD-FRQNRYFYHLSGVDIPASAILFNCSTDKLTLFLPNIDEEDVIWSGMPLSLDEAM 171
Query 65 SRYAVTDAIAVEAISDVEAELQRRGVRTVHVLKGVNSDSGRPVAPPKALNSFSSFTVDCS 124
+ + +A+ + + ELQ + T + + + R + P D +
Sbjct 172 RVFDIDEALYISDLGKKFKELQDFAIFTTDLDNVHDENIARSLIPS-----------DPN 220
Query 125 ALYDILVECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYVGFVG 184
Y + E R K E E + AC S ++H V + + E Q +A F+ + G
Sbjct 221 FFY-AMDETRAIKDWYEIESIRKACQISDKSHLAVMSALPIELNELQIQAEFEYHATRQG 279
Query 185 GARHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPS 244
G R + YD ICC+G LHY N IK +L D G ++ Y +DIT +P+
Sbjct 280 G-RSLGYDPICCSGPACGTLHYVK----NSEDIKGKHSILIDAGAEWRQYTSDITRCFPT 334
Query 245 NGIFTETQKAIYDAVLDAQKSVIERMRPGVEWTELHRLAELVILKHL 291
+G FT + +Y+ VLD Q +ER++PG +W +LH L V++KH
Sbjct 335 SGKFTAEHREVYETVLDMQNQAMERIKPGAKWDDLHALTHKVLIKHF 381
> ath:AT1G09300 metallopeptidase M24 family protein; K01262 Xaa-Pro
aminopeptidase [EC:3.4.11.9]
Length=462
Score = 100 bits (248), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 75/277 (27%), Positives = 127/277 (45%), Gaps = 23/277 (8%)
Query 10 AFRQEAFFQHLVGVNEPDIYAAFVLNSHELLLFIPKIPADALRFMGPSKDADFYRSRYAV 69
FRQ+A + +L G +P A + + L +F+P+ + + G D +
Sbjct 60 TFRQDADYLYLTGCQQPG-GVAVLSDERGLCMFMPESTPKDIAWEGEVAGVDAASEVFKA 118
Query 70 TDAIAVEAISDVEAELQRRGVRTVHVLKGVNSDSGRPVAPPKALNSFSSFTVDCSALYDI 129
A + + ++ +++ R + H V S S R NS S V L +
Sbjct 119 DQAYPISKLPEILSDMIRHSSKVFH---NVQSASQRYTNLDDFQNSASLGKV--KTLSSL 173
Query 130 LVECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQA---EAIFKAYVGF---V 183
E RL K+P E + + + + Q + M+ + E I A V + V
Sbjct 174 THELRLIKSPAELKLMRESASIACQG-------LLKTMLHSKGFPDEGILSAQVEYECRV 226
Query 184 GGARHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYP 243
GA+ +A++ + G NAS++HY ND IKD DL+L DMG + +GY +D+T T+P
Sbjct 227 RGAQRMAFNPVVGGGSNASVIHYSR----NDQRIKDGDLVLMDMGCELHGYVSDLTRTWP 282
Query 244 SNGIFTETQKAIYDAVLDAQKSVIERMRPGVEWTELH 280
G F+ Q+ +YD +L K I++ +PG +L+
Sbjct 283 PCGKFSSVQEELYDLILQTNKECIKQCKPGTTIRQLN 319
> eco:b2908 pepP, ECK2903, JW2876; proline aminopeptidase P II
(EC:3.4.11.9); K01262 Xaa-Pro aminopeptidase [EC:3.4.11.9]
Length=441
Score = 99.4 bits (246), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 58/155 (37%), Positives = 84/155 (54%), Gaps = 13/155 (8%)
Query 129 ILVECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGM----VEGQAEAIFKAYVGFVG 184
++ E RL K+P E L A ++ AH R GM +EG+ F +
Sbjct 169 VVHEMRLFKSPEEIAVLRRAGEITAMAHTRAMEKCRPGMFEYHLEGEIHHEFNRH----- 223
Query 185 GARHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPS 244
GAR+ +Y+ I +GEN ILHY N+ ++D DL+L D G +Y GYA DIT T+P
Sbjct 224 GARYPSYNTIVGSGENGCILHYTE----NECEMRDGDLVLIDAGCEYKGYAGDITRTFPV 279
Query 245 NGIFTETQKAIYDAVLDAQKSVIERMRPGVEWTEL 279
NG FT+ Q+ IYD VL++ ++ + RPG E+
Sbjct 280 NGKFTQAQREIYDIVLESLETSLRLYRPGTSILEV 314
> xla:446780 xpnpep3, MGC80423; X-prolyl aminopeptidase (aminopeptidase
P) 3, putative (EC:3.4.11.9); K01262 Xaa-Pro aminopeptidase
[EC:3.4.11.9]
Length=502
Score = 87.8 bits (216), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 89/298 (29%), Positives = 128/298 (42%), Gaps = 31/298 (10%)
Query 11 FRQEAFFQHLVGVNEPD------IYAAFVLNSHELLLFIPKIPADALRFMGPSKDADFYR 64
F Q F +L G EPD + SH L++P+ + GP D
Sbjct 109 FHQNNDFLYLCGFLEPDSILLLQSQSRQSSLSHTAKLYVPRRDPGRELWDGPRSGTDGAV 168
Query 65 SRYAVTDAIAVEAISDVEAELQRRGVR-----TVHVLKGVNSDSGRPVAPPKALNSFSSF 119
S V +A E V L GV T V +++ S +P L F S
Sbjct 169 SLTGVDEAFTTEEFKHVLPRLYDEGVTVWYDCTTPVHPALHTSSLQP------LVEFRSR 222
Query 120 TVD-CSALYDILVECRLRKTPLEAEYLAAACLCSSQAHCFV-ARNIRSGMVEGQAEAIFK 177
+ + +L ++ + RL K+ E E + A SSQA F+ + R VE EA
Sbjct 223 SKNRIRSLRHLVQQLRLVKSQAEVELMKKAGYISSQA--FIETMSCRKAPVE---EAFLY 277
Query 178 AYVGF---VGGARHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGY 234
A F GA +AY + G A+ LHY N+ IK +++L D G + + Y
Sbjct 278 AKFDFECRARGADILAYPPVVAGGNRANTLHYVK----NNQIIKSGEMVLLDGGCEASCY 333
Query 235 ATDITFTYPSNGIFTETQKAIYDAVLDAQKSVIERMRPGVEWTELHRLAELVILKHLK 292
+DIT T+P NG FT Q+A+Y AVLD QKS + PG ++ +I + LK
Sbjct 334 VSDITRTWPVNGRFTAPQEALYQAVLDVQKSCLRLCYPGTSLENIYSHMLAMIARKLK 391
> hsa:63929 XPNPEP3, APP3, NPHPL1; X-prolyl aminopeptidase (aminopeptidase
P) 3, putative (EC:3.4.11.9); K01262 Xaa-Pro aminopeptidase
[EC:3.4.11.9]
Length=507
Score = 86.3 bits (212), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 79/308 (25%), Positives = 134/308 (43%), Gaps = 34/308 (11%)
Query 10 AFRQEAFFQHLVGVNEPDIYAAFV------LNSHELLLFIPKIPADALRFMGPSKDADFY 63
F Q+ F +L G EPD L SH+ +LF+P+ + GP D
Sbjct 114 TFHQDNNFLYLCGFQEPDSILVLQSLPGKQLPSHKAILFVPRRDPSRELWDGPRSGTDGA 173
Query 64 RSRYAVTDAIAVEAISDVEAELQRRG-------VRTVHVLKGVNSDSGRPVAPPKALNSF 116
+ V +A +E + +++ +R H ++SD +P+ KA +
Sbjct 174 IALTGVDEAYTLEEFQHLLPKMKAETNMVWYDWMRPSHA--QLHSDYMQPLTEAKAKSKN 231
Query 117 SSFTVDCSALYDILVECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIF 176
+ ++ RL K+P E E + A +SQA ++ + E A F
Sbjct 232 K-----VRGVQQLIQRLRLIKSPAEIERMQIAGKLTSQAFIETMFTSKAPVEEAFLYAKF 286
Query 177 KAYVGFVGGARHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYAT 236
+ + GA +AY + G ++ LHY N+ IKD +++L D G + + Y +
Sbjct 287 E-FECRARGADILAYPPVVAGGNRSNTLHYVK----NNQLIKDGEMVLLDGGCESSCYVS 341
Query 237 DITFTYPSNGIFTETQKAIYDAVLDAQKSVIERMRPGVEWTELH---------RLAELVI 287
DIT T+P NG FT Q +Y+AVL+ Q+ + PG ++ +L +L I
Sbjct 342 DITRTWPVNGRFTAPQAELYEAVLEIQRDCLALCFPGTSLENIYSMMLTLIGQKLKDLGI 401
Query 288 LKHLKVRN 295
+K++K N
Sbjct 402 MKNIKENN 409
> dre:573360 xpnpep3, MGC76905, zgc:76905; X-prolyl aminopeptidase
(aminopeptidase P) 3, putative (EC:3.4.11.9); K01262 Xaa-Pro
aminopeptidase [EC:3.4.11.9]
Length=510
Score = 78.6 bits (192), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 75/290 (25%), Positives = 127/290 (43%), Gaps = 25/290 (8%)
Query 11 FRQEAFFQHLVGVNEPDIYAAFVL----NSHELLLFIPKI-PADALRFMGPSKDADFYRS 65
F Q F +L G+ EPD +A V+ + +LF+P+ PA L + GP D +
Sbjct 125 FHQNQDFLYLTGITEPD--SALVMYGSSKPDQAVLFVPRRDPAREL-WDGPRSGKDGAAA 181
Query 66 RYAVTDAIAVEAISDVEAELQRRGV---RTVHVLKGVNSDSGRPVAPPKALNSFSSFTVD 122
+ + E + V ++ V + + ++ RP+ L
Sbjct 182 LTGLERVHSTEELGVVLKSIKGGTVWYDNSQPCHQRLHQTHVRPLLEGGQL--------- 232
Query 123 CSALYDILVECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYVGF 182
+L + R K+P E + A ++QA R + E A F Y
Sbjct 233 VKSLRPLTHSLRAIKSPAEVALMKEAGRITAQAFKKTMAMSRGNIDEAVLYAKFD-YECR 291
Query 183 VGGARHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTY 242
GA +AY + G A+ LHY + N+ +KD +++L D G +Y GY +DIT T+
Sbjct 292 AHGANFLAYPPVVAGGNRANTLHYIN----NNQIVKDGEMVLLDGGCEYFGYVSDITRTW 347
Query 243 PSNGIFTETQKAIYDAVLDAQKSVIERMRPGVEWTELHRLAELVILKHLK 292
P NG F+ Q+ +Y+AVL+ Q + + + PGV ++ ++ + LK
Sbjct 348 PVNGKFSAAQRELYEAVLEVQLACLSQCSPGVSLDYIYSTMLTLLARQLK 397
> sce:YER078C ICP55; Icp55p (EC:3.4.-.-); K01262 Xaa-Pro aminopeptidase
[EC:3.4.11.9]
Length=511
Score = 66.2 bits (160), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 53/197 (26%), Positives = 92/197 (46%), Gaps = 20/197 (10%)
Query 111 KALNSFSSFTVDCSALYDILVECRLRKTPLEAEYLAAACLCSSQA--HCFVARNIRSGMV 168
++LNS ++ T+ + + E R K+P E + A S ++ F R +
Sbjct 221 RSLNSIANKTI--KPISKRIAEFRKIKSPQELRIMRRAGQISGRSFNQAFAKRFRNERTL 278
Query 169 EGQAEAIFKAYVGFVGGARHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMG 228
+ F Y GG AY + G N+ +HY ND + D++++L D
Sbjct 279 DS-----FLHYKFISGGCDKDAYIPVVATGSNSLCIHYTR----NDDVMFDDEMVLVDAA 329
Query 229 GDYNGYATDITFTYPSNGIFTETQKAIYDAVLDAQKSVIERMRPGVEWTELHRLAELVI- 287
G GY DI+ T+P++G FT+ Q+ +Y+AVL+ Q+ I+ + ++ LH + E I
Sbjct 330 GSLGGYCADISRTWPNSGKFTDAQRDLYEAVLNVQRDCIKLCKASNNYS-LHDIHEKSIT 388
Query 288 -----LKHLKVRNDTLW 299
LK+L + + W
Sbjct 389 LMKQELKNLGIDKVSGW 405
> mmu:321003 Xpnpep3, APP3, E430012M05Rik; X-prolyl aminopeptidase
(aminopeptidase P) 3, putative (EC:3.4.11.9); K01262 Xaa-Pro
aminopeptidase [EC:3.4.11.9]
Length=386
Score = 65.1 bits (157), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 66/270 (24%), Positives = 115/270 (42%), Gaps = 25/270 (9%)
Query 10 AFRQEAFFQHLVGVNEPDI------YAAFVLNSHELLLFIPKIPADALRFMGPSKDADFY 63
F Q+ F +L G EPD ++ L SH+ +LF+P+ + GP D
Sbjct 114 TFHQDNNFLYLCGFQEPDSILVLQSFSGKQLPSHKAMLFVPRRDPGRELWDGPRSGTDGA 173
Query 64 RSRYAVTDAIAVEAISDVEAELQRRG-------VRTVHVLKGVNSDSGRPVAPPKALNSF 116
+ V +A +E + +L+ ++ H ++SD +P+ KA +
Sbjct 174 IALTGVDEAYPLEEFQHLLPKLRAETNMVWYDWMKPSHA--QLHSDYMQPLTEAKARSKN 231
Query 117 SSFTVDCSALYDILVECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIF 176
+V ++ RL K+P E + + A +S+A ++ + E A F
Sbjct 232 KVRSVQ-----QLIQRLRLVKSPSEIKRMQIAGKLTSEAFIETMFASKAPIDEAFLYAKF 286
Query 177 KAYVGFVGGARHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYAT 236
+ + GA +AY + G ++ LHY N+ IKD +++L D G + + Y +
Sbjct 287 E-FECRARGADILAYPPVVAGGNRSNTLHYVK----NNQLIKDGEMVLLDGGCESSCYVS 341
Query 237 DITFTYPSNGIFTETQKAIYDAVLDAQKSV 266
DIT T+P NG E I A+ S+
Sbjct 342 DITRTWPVNGRLLENTALIMLAITSGWMSM 371
> eco:b2385 ypdF, ECK2381, JW2382; Xaa-Pro aminopeptidase; K08326
aminopeptidase [EC:3.4.11.-]
Length=361
Score = 63.2 bits (152), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 49/169 (28%), Positives = 78/169 (46%), Gaps = 12/169 (7%)
Query 123 CSALYDILVECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYVGF 182
SA D+L R KTP E E + AC + + + R I++GM E + A + ++
Sbjct 117 VSATPDVL---RQIKTPEEVEKIRLACGIADRGAEHIRRFIQAGMSEREIAAELEWFM-R 172
Query 183 VGGARHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTY 242
GA ++D I +G ++ H G+ +D + + + D G Y GY +D+T T
Sbjct 173 QQGAEKASFDTIVASGWRGALPH----GKASDKIVAAGEFVTLDFGALYQGYCSDMTRTL 228
Query 243 PSNGIFTETQK----AIYDAVLDAQKSVIERMRPGVEWTELHRLAELVI 287
NG + +Y VL AQ + I +RPGV ++ A VI
Sbjct 229 LVNGEGVSAESHLLFNVYQIVLQAQLAAISAIRPGVRCQQVDDAARRVI 277
> eco:b3847 pepQ, ECK3839, JW3823; proline dipeptidase (EC:3.4.13.9);
K01271 Xaa-Pro dipeptidase [EC:3.4.13.9]
Length=443
Score = 51.6 bits (122), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 49/188 (26%), Positives = 77/188 (40%), Gaps = 21/188 (11%)
Query 109 PPKALN-SFSSFTVDCSALYDILVECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGM 167
P +AL + ++ + D L R KT E + A + H RSGM
Sbjct 133 PERALQLGIEASNINPKGVIDYLHYYRSFKTEYELACMREAQKMAVNGHRAAEEAFRSGM 192
Query 168 VEGQAEAIFKAYVGFVGGARH----VAYDCICCAGENASILHYGHAGRPNDGSIKDNDLL 223
E F + ++ H V Y I E+A++LHY ++
Sbjct 193 SE------FDINIAYLTATGHRDTDVPYSNIVALNEHAAVLHYTKLDHQAPEEMRS---F 243
Query 224 LFDMGGDYNGYATDITFTYPSNGIFTETQKAIYDAVLDAQKSVIERMRPGVEWTELH--- 280
L D G +YNGYA D+T T+ + Q + V D Q ++I M+ GV + + H
Sbjct 244 LLDAGAEYNGYAADLTRTWSAKSDNDYAQ--LVKDVNDEQLALIATMKAGVSYVDYHIQF 301
Query 281 --RLAELV 286
R+A+L+
Sbjct 302 HQRIAKLL 309
> cel:R119.2 hypothetical protein; K01262 Xaa-Pro aminopeptidase
[EC:3.4.11.9]
Length=362
Score = 50.1 bits (118), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 36/137 (26%), Positives = 65/137 (47%), Gaps = 11/137 (8%)
Query 134 RLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYVGFVG---GARHVA 190
R+ K+P E + C +Q ++ SG + E + F G G+ A
Sbjct 98 RVIKSPSEMSSMRDVCNVGAQ----TMSSMISGSRDLHNENAICGLLEFEGRRRGSEMQA 153
Query 191 YDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTE 250
Y + G A+ +HY A ND + + +L D G D NGY +D+T +P +G +++
Sbjct 154 YPPVIAGGVRANTIHYLDAN--ND--LNPRECVLVDAGCDLNGYVSDVTRCFPISGFWSD 209
Query 251 TQKAIYDAVLDAQKSVI 267
Q ++Y+A+L + ++
Sbjct 210 AQLSLYEALLYVHEELL 226
> dre:100333156 peptidase-like
Length=611
Score = 45.1 bits (105), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 35/117 (29%), Positives = 51/117 (43%), Gaps = 5/117 (4%)
Query 176 FKAYVG--FVGGARHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNG 233
F+A VG F + +++D I AG N +I+HY N + +L L D G Y
Sbjct 362 FRASVGQDFQNPLKDISFDTISGAGPNGAIIHYRVTTETNR-KLSSGELFLIDSGAQYVN 420
Query 234 YATDITFTYPSNGIFTETQKAIYDAVLDAQKSV-IERMRPGVEWTELHRLAELVILK 289
TDIT T + G + QK + VL + R G +L LA + + K
Sbjct 421 GTTDITRTV-AIGAVPQEQKRFFTLVLKGMIGISTARFPKGTRGCDLDPLARIALWK 476
> xla:414716 hypothetical protein MGC83093; K01262 Xaa-Pro aminopeptidase
[EC:3.4.11.9]
Length=621
Score = 42.0 bits (97), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 44/174 (25%), Positives = 72/174 (41%), Gaps = 23/174 (13%)
Query 108 APPKALNSFSSFTVDCSALYDILVECRLRKTPLEAEYLAAACLCSSQAHC----FVARNI 163
A PK S ++ C A + K P+E E + A + + A C ++ + I
Sbjct 299 AIPKTHRLLSQYSPICLA--------KAVKNPVETEGMRRAHVKDAVALCELFNWLEKEI 350
Query 164 RSGMVE-----GQAEAIFKAYVGFVGGARHVAYDCICCAGENASILHYGHAGRPNDGSIK 218
G V +AE + V FV +++ I +G NA+I+HY N +
Sbjct 351 PKGTVTEISAADKAEEFRRQQVDFV----ELSFATISSSGPNAAIIHYKPVPETNR-QLS 405
Query 219 DNDLLLFDMGGDYNGYATDITFTYPSNGIFTETQKAIYDAVLDAQKSVIERMRP 272
N++ L D G + TD+T T G T+ +K + VL +V + P
Sbjct 406 ANEIFLLDSGAQFKDGTTDVTRTL-HFGTPTDYEKECFTYVLQGHIAVSSAVFP 458
> tgo:TGME49_048850 methionine aminopeptidase, putative (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=416
Score = 40.8 bits (94), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 31/154 (20%), Positives = 63/154 (40%), Gaps = 8/154 (5%)
Query 141 EAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYV----GFVGGARHVAYDCICC 196
E + L CL +A + ++ G+ + +A A++ G+ + + CC
Sbjct 159 EIQRLRETCLLGRRALDYAHSLVKPGVTTEEIDAKVHAFIVDNGGYPSPLNYQQFPKSCC 218
Query 197 AGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTETQKAIY 256
N I H G P+ ++D D++ D+ + G D+ TY E K +
Sbjct 219 TSVNEVICH----GIPDFRPLQDGDIVNIDITVFFKGMHGDLNETYCVGDNVDEDSKRLI 274
Query 257 DAVLDAQKSVIERMRPGVEWTELHRLAELVILKH 290
+ +++ RPG+ + ++ R+ V K+
Sbjct 275 KGAYECLMEAVKQCRPGMMYRDVGRIVSDVADKY 308
> cpv:cgd4_2910 aminopeptidase
Length=694
Score = 40.0 bits (92), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 26/63 (41%), Positives = 33/63 (52%), Gaps = 6/63 (9%)
Query 190 AYDCICCAGENASILHYGHAGRPN--DGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGI 247
++D I GEN +I+HY RP + SI DL L D GG Y+ TD+T T GI
Sbjct 433 SFDTISSIGENGAIIHY----RPEKENSSIIKPDLYLCDSGGQYHTGTTDVTRTLFLFGI 488
Query 248 FTE 250
E
Sbjct 489 GEE 491
> dre:100333662 methionyl aminopeptidase 1-like
Length=249
Score = 37.4 bits (85), Expect = 0.065, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 39/85 (45%), Gaps = 5/85 (5%)
Query 195 CCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTETQKA 254
CC N I H G P+ +++ D+L D+ +NGY D+ T+ G E K
Sbjct 55 CCTSVNEVICH----GIPDRRHLQEGDILNIDITVYHNGYHGDLNETF-FVGEVDEGAKR 109
Query 255 IYDAVLDAQKSVIERMRPGVEWTEL 279
+ + I+ ++PG+ + EL
Sbjct 110 LVQTTYECLMQAIDSVKPGIRYREL 134
> dre:503783 metap1, im:7047238, wu:fc84e12, zgc:110093; methionyl
aminopeptidase 1 (EC:3.4.11.18); K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=386
Score = 37.4 bits (85), Expect = 0.070, Method: Compositional matrix adjust.
Identities = 23/90 (25%), Positives = 41/90 (45%), Gaps = 5/90 (5%)
Query 195 CCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTETQKA 254
CC N I H G P+ +++ D+L D+ +NGY D+ T+ G E K
Sbjct 192 CCTSVNEVICH----GIPDRRHLQEGDILNIDITVYHNGYHGDLNETF-FVGEVDEGAKR 246
Query 255 IYDAVLDAQKSVIERMRPGVEWTELHRLAE 284
+ + I+ ++PG+ + EL + +
Sbjct 247 LVQTTYECLMQAIDSVKPGIRYRELGNITQ 276
> hsa:7512 XPNPEP2, APP2; X-prolyl aminopeptidase (aminopeptidase
P) 2, membrane-bound (EC:3.4.11.9); K14208 Xaa-Pro aminopeptidase
2 [EC:3.4.11.9]
Length=674
Score = 35.8 bits (81), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 31/108 (28%), Positives = 49/108 (45%), Gaps = 9/108 (8%)
Query 157 CFVARNIRSGMVE--GQAEAI--FKAYVGFVGGARHVAYDCICCAGENASILHYGHAGRP 212
++ +N+ G V+ AE + F+ F G +++ I +G NA++ HY
Sbjct 381 VWLEKNVPKGTVDEFSGAEIVDKFRGEEQFSSGP---SFETISASGLNAALAHYSPTKEL 437
Query 213 NDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTETQKAIYDAVL 260
N + +++ L D GG Y TDIT T G + QK Y VL
Sbjct 438 NR-KLSSDEMYLLDSGGQYWDGTTDITRTV-HWGTPSAFQKEAYTRVL 483
> ath:AT2G45240 MAP1A; MAP1A (METHIONINE AMINOPEPTIDASE 1A); aminopeptidase/
metalloexopeptidase; K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=398
Score = 35.8 bits (81), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 32/147 (21%), Positives = 59/147 (40%), Gaps = 9/147 (6%)
Query 137 KTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFK----AYVGFVGGARHVAYD 192
KTP + + + C + + AR I G+ + + + A G+ + +
Sbjct 142 KTPEQIQRMRETCKIAREVLDAAARVIHPGVTTDEIDRVVHEATIAAGGYPSPLNYYFFP 201
Query 193 CICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTETQ 252
CC N I H G P+ ++D D++ D+ Y G D+ TY G E
Sbjct 202 KSCCTSVNEVICH----GIPDARKLEDGDIVNVDVTVCYKGCHGDLNETY-FVGNVDEAS 256
Query 253 KAIYDAVLDAQKSVIERMRPGVEWTEL 279
+ + + + I ++PGV + E+
Sbjct 257 RQLVKCTYECLEKAIAIVKPGVRFREI 283
> dre:394007 xpnpep2, MGC63528, zgc:63528; X-prolyl aminopeptidase
(aminopeptidase P) 2, membrane-bound (EC:3.4.11.9); K14208
Xaa-Pro aminopeptidase 2 [EC:3.4.11.9]
Length=702
Score = 35.8 bits (81), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 50/185 (27%), Positives = 79/185 (42%), Gaps = 14/185 (7%)
Query 80 DVEAELQRRGVRTVHVLKGVNSDSGRPVAPP-KALNSFSSFTVDCSALYDILVECRLRKT 138
D+++ LQR VR + N + P K L S S + A+ D + E R+ K
Sbjct 335 DLQSYLQRPNVRVWVGTEYTNQALYELITPEDKLLTSTYSPVLTTKAVKD-MTEQRILK- 392
Query 139 PLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYV-GFVGGARHVAYDCICCA 197
EA A + Q ++ + + G A++ +R +++ I +
Sbjct 393 --EAHVRDAVAVM--QLLLWLEKKVPEGAETEITAALYADQCRSKQKNSRGPSFETISAS 448
Query 198 GENASILHYGHAGRPNDGSIK--DNDLLLFDMGGDYNGYATDITFTYPSNGIFTETQKAI 255
G NA++ HY + ND + K +++ L D GG Y TDIT T G T+ QK
Sbjct 449 GPNAALAHYSPS---NDTARKLTVDEMYLVDSGGQYLDGTTDITRTV-HWGKPTDFQKEA 504
Query 256 YDAVL 260
Y VL
Sbjct 505 YTRVL 509
> cel:W03G9.4 app-1; AminoPeptidase P family member (app-1); K01262
Xaa-Pro aminopeptidase [EC:3.4.11.9]
Length=616
Score = 35.0 bits (79), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 26/53 (49%), Gaps = 0/53 (0%)
Query 189 VAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFT 241
+++D I G++A++ HY G + N + L D G Y TD+T T
Sbjct 376 LSFDTISAVGDHAALPHYKPLGESGNRKAAANQVFLLDSGAHYGDGTTDVTRT 428
> dre:406253 xpnpep1, MGC56366, fa02e02, wu:fa02e02, zgc:56366,
zgc:77772; X-prolyl aminopeptidase (aminopeptidase P) 1, soluble
(EC:3.4.11.9); K01262 Xaa-Pro aminopeptidase [EC:3.4.11.9]
Length=620
Score = 35.0 bits (79), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 34/128 (26%), Positives = 55/128 (42%), Gaps = 19/128 (14%)
Query 147 AACLCSSQAHCFVARNIRSGMVE-----GQAEAIFKAYVGFVGGARHVAYDCICCAGENA 201
A LC + ++ + I G V +AE + FVG +++ I G N
Sbjct 338 AVALC--ELFAWLEKEIPKGTVTEISAADKAEELRSQQKEFVG----LSFPTISSVGPNG 391
Query 202 SILHYGHAGRP---NDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTETQKAIYDA 258
+I+HY RP + ++ N++ L D G Y TD+T T G +E +K +
Sbjct 392 AIIHY----RPLPETNRTLSLNEVYLIDSGAQYTDGTTDVTRTV-HFGTPSEYEKECFTY 446
Query 259 VLDAQKSV 266
VL +V
Sbjct 447 VLKGHIAV 454
> cpv:cgd1_2700 methionine aminopeptidase with MYND finger at
N-terminus ; K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=407
Score = 33.5 bits (75), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 36/160 (22%), Positives = 66/160 (41%), Gaps = 15/160 (9%)
Query 138 TPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYVGFV-------GGARHVA 190
T E E L C +A A I+ G+ +AI +A F+ +
Sbjct 151 TAEEIELLRECCKIGREALDIAASMIKPGVT---TDAIDEAVHNFIISKNSYPSPLNYWE 207
Query 191 YDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTE 250
+ CC N I H G P+ +++ D++ D+ Y G +D+ T+P + +
Sbjct 208 FPKSCCTSVNEIICH----GIPDFRPLEEGDIVNVDISVYYKGVHSDLNETFPVGKVDEK 263
Query 251 TQKAIYDAVLDAQKSVIERMRPGVEWTELHRLAELVILKH 290
+ K + A ++S I+ +PG + E+ L + + K
Sbjct 264 SMKLMKVAYQCLEES-IKICKPGTMYREIGNLIQSICDKQ 302
> mmu:170745 Xpnpep2, 9030008G12Rik, mAPP; X-prolyl aminopeptidase
(aminopeptidase P) 2, membrane-bound (EC:3.4.11.9); K14208
Xaa-Pro aminopeptidase 2 [EC:3.4.11.9]
Length=674
Score = 33.5 bits (75), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 32/111 (28%), Positives = 49/111 (44%), Gaps = 9/111 (8%)
Query 154 QAHCFVARNIRSGMVE--GQAEAI--FKAYVGFVGGARHVAYDCICCAGENASILHYGHA 209
Q ++ +N+ G V+ AE I + F G +++ I +G NA++ HY
Sbjct 378 QYLVWLEKNVPKGTVDEFSGAEYIDELRRNENFSSGP---SFETISASGLNAALAHYSPT 434
Query 210 GRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTETQKAIYDAVL 260
+ + +++ L D GG Y TDIT T G T QK Y VL
Sbjct 435 KELHR-KLSSDEMYLVDSGGQYWDGTTDITRTV-HWGTPTAFQKEAYTRVL 483
> bbo:BBOV_IV006760 23.m06237; methionine aminopeptidase (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=376
Score = 33.5 bits (75), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 31/156 (19%), Positives = 66/156 (42%), Gaps = 15/156 (9%)
Query 138 TPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYV----GFVGGARHVAYDC 193
TP + + + A + +A F A I G+ + + ++ + + +
Sbjct 123 TPEQIKLIRKASILGRKALDFAASLIAPGVTTDEIDTKVHDFIIQHNAYPSPLNYYGFPK 182
Query 194 ICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTETQ- 252
C N + H G P+ +KD D++ D+ NG +D+ TY + +++
Sbjct 183 SLCTSVNEVVCH----GIPDKRPLKDGDIINIDISVYLNGVHSDLNATYFVGEVDEDSRR 238
Query 253 --KAIYDAVLDAQKSVIERMRPGVEWTELHRLAELV 286
K Y A+++A I++ +PG+ + E+ + V
Sbjct 239 LVKGTYMALMEA----IKQCKPGMYYREIGNIINKV 270
> ath:AT2G27480 calcium ion binding; K13448 calcium-binding protein
CML
Length=228
Score = 33.1 bits (74), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 33/61 (54%), Gaps = 3/61 (4%)
Query 3 SADCNKAAFRQEAFFQHLVGVNEPDIYAAFVLNSHELLLFIPKIPADALRFMGPSKDADF 62
SAD N++ F +E+ + + ++ Y + LLLFI KIP D+L +GP + +
Sbjct 63 SADRNRSGFLEESELRQALSLSG---YDGISNRTIRLLLFIYKIPVDSLLRLGPKEYVEL 119
Query 63 Y 63
+
Sbjct 120 W 120
> ath:AT3G05350 aminopeptidase/ hydrolase; K01262 Xaa-Pro aminopeptidase
[EC:3.4.11.9]
Length=710
Score = 32.7 bits (73), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 30/67 (44%), Gaps = 11/67 (16%)
Query 185 GARHVAYDCICCAGENASILHYGHAGRPNDGS---IKDNDLLLFDMGGDYNGYATDIT-- 239
G ++D I +G N +I+HY +P S + L L D G Y TDIT
Sbjct 466 GFMDTSFDTISGSGANGAIIHY----KPEPESCSRVDPQKLFLLDSGAQYVDGTTDITRT 521
Query 240 --FTYPS 244
F+ PS
Sbjct 522 VHFSEPS 528
> tpv:TP01_0224 hypothetical protein; K03564 peroxiredoxin Q/BCP
[EC:1.11.1.15]
Length=186
Score = 32.7 bits (73), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 26/93 (27%), Positives = 37/93 (39%), Gaps = 19/93 (20%)
Query 223 LLFDMGGDYNGYATDITFTYPSNGI---------FTETQKAIYDAVLDAQKSVIERMRPG 273
LL ++ DY G + F +PS G FT KA D + P
Sbjct 73 LLGELPPDYKGL---VLFLFPSAGTPGCTKQACNFTSNHKAFKDKGYEVYGLTSSERTPA 129
Query 274 VEWTELHRLA-------ELVILKHLKVRNDTLW 299
+WTE H+L+ EL ++ HL R L+
Sbjct 130 GKWTEKHKLSFTTLVDKELKLVDHLDCRKLGLF 162
> bbo:BBOV_III008370 17.m07732; metallopeptidase M24 family protein;
K01262 Xaa-Pro aminopeptidase [EC:3.4.11.9]
Length=624
Score = 32.3 bits (72), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 32/134 (23%), Positives = 50/134 (37%), Gaps = 10/134 (7%)
Query 130 LVECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYVGFVGGARHV 189
+ E ++ AE+ A F A + G + Q A R +
Sbjct 322 MTEAHIQDAIALAEFFAKVENMKQDGTLFTADELILGSMSSQCRADMP-------DNRGI 374
Query 190 AYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFT 249
++ I G N +++HY A I+ + L D GG Y G TD+T T G +
Sbjct 375 SFHPISSIGSNCAVVHY-RATEEIKAKIEPK-IYLLDSGGQYPGGTTDVTRTI-HFGTPS 431
Query 250 ETQKAIYDAVLDAQ 263
+ +K Y VL
Sbjct 432 DEEKEAYTQVLKGH 445
> ath:AT4G37040 MAP1D; MAP1D (METHIONINE AMINOPEPTIDASE 1D); aminopeptidase/
metalloexopeptidase; K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=350
Score = 32.0 bits (71), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 42/95 (44%), Gaps = 5/95 (5%)
Query 196 CAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTETQKAI 255
C N I H G P+ ++D D++ D+ NGY D + T+ G E K +
Sbjct 171 CTSVNECICH----GIPDSRPLEDGDIINIDVTVYLNGYHGDTSATFFC-GNVDEKAKKL 225
Query 256 YDAVLDAQKSVIERMRPGVEWTELHRLAELVILKH 290
+ ++ I PGVE+ ++ ++ + KH
Sbjct 226 VEVTKESLDKAISICGPGVEYKKIGKVIHDLADKH 260
> pfa:PF14_0517 peptidase, putative; K01262 Xaa-Pro aminopeptidase
[EC:3.4.11.9]
Length=764
Score = 32.0 bits (71), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 36/71 (50%), Gaps = 3/71 (4%)
Query 190 AYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFT 249
++ I +G NA+++HY + N +IK + L D GG Y TD+T T G T
Sbjct 523 SFSTISASGPNAAVIHYECTDKTN-ATIKP-AIYLLDSGGQYLHGTTDVTRT-THFGEPT 579
Query 250 ETQKAIYDAVL 260
+K IY VL
Sbjct 580 AEEKRIYTLVL 590
> mmu:170750 Xpnpep1, D230045I08Rik; X-prolyl aminopeptidase (aminopeptidase
P) 1, soluble (EC:3.4.11.9); K01262 Xaa-Pro aminopeptidase
[EC:3.4.11.9]
Length=623
Score = 31.2 bits (69), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 21/78 (26%), Positives = 35/78 (44%), Gaps = 2/78 (2%)
Query 189 VAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIF 248
+++ I G N +I+HY N ++ +++ L D G Y TD+T T G
Sbjct 379 LSFPTISSTGPNGAIIHYAPVPETNR-TLSLDEVYLIDSGAQYKDGTTDVTRTM-HFGTP 436
Query 249 TETQKAIYDAVLDAQKSV 266
T +K + VL +V
Sbjct 437 TAYEKECFTYVLKGHIAV 454
Lambda K H
0.322 0.136 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 11902145332
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40