bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1017_orf2
Length=156
Score E
Sequences producing significant alignments: (Bits) Value
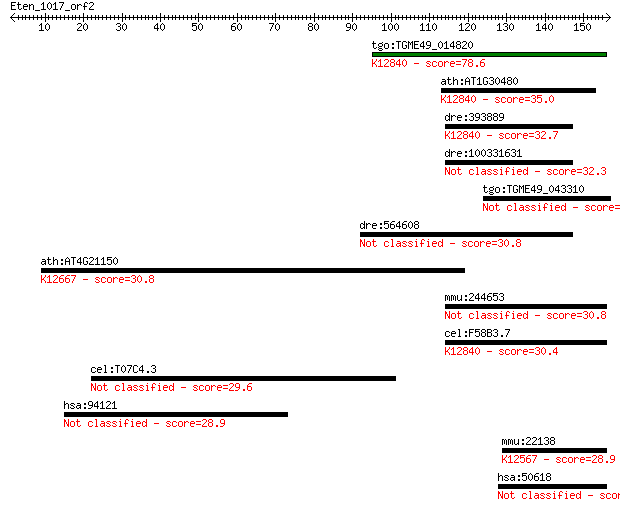
tgo:TGME49_014820 DNA repair enzyme, putative ; K12840 splicin... 78.6 7e-15
ath:AT1G30480 DRT111; DRT111; nucleic acid binding / nucleotid... 35.0 0.092
dre:393889 rbm17, MGC55840, zgc:55840; RNA binding motif prote... 32.7 0.53
dre:100331631 RNA binding motif protein 17-like 32.3 0.56
tgo:TGME49_043310 hypothetical protein 32.0 0.79
dre:564608 tmem198b, MGC112212, tmem198, zgc:112212; transmemb... 30.8 1.6
ath:AT4G21150 HAP6; HAP6 (HAPLESS 6); dolichyl-diphosphooligos... 30.8 1.7
mmu:244653 Hydin, 1700034M11Rik, 4930545D19Rik, 4932703P14, A8... 30.8 2.0
cel:F58B3.7 hypothetical protein; K12840 splicing factor 45 30.4
cel:T07C4.3 hypothetical protein 29.6 4.7
hsa:94121 SYTL4, DKFZp451P0116, FLJ40960, SLP4; synaptotagmin-... 28.9 7.3
mmu:22138 Ttn, 1100001C23Rik, 2310036G12Rik, 2310057K23Rik, 23... 28.9 7.4
hsa:50618 ITSN2, KIAA1256, PRO2015, SH3D1B, SH3P18, SWA, SWAP;... 28.5 9.2
> tgo:TGME49_014820 DNA repair enzyme, putative ; K12840 splicing
factor 45
Length=466
Score = 78.6 bits (192), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 34/61 (55%), Positives = 45/61 (73%), Gaps = 0/61 (0%)
Query 95 GSSSPLDPTTWVPVGLWPQEYDPSKPNDYTAIHKEKMRQQQREELERLRQQELEKQKHDE 154
G + + +V +G EYDP PNDY+AI++EK+R+QQREELERLRQQEL+KQ+ D
Sbjct 196 GGDKETESSGFVSIGQMKDEYDPGTPNDYSAIYREKIRKQQREELERLRQQELDKQRRDR 255
Query 155 E 155
E
Sbjct 256 E 256
> ath:AT1G30480 DRT111; DRT111; nucleic acid binding / nucleotide
binding; K12840 splicing factor 45
Length=387
Score = 35.0 bits (79), Expect = 0.092, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 26/40 (65%), Gaps = 4/40 (10%)
Query 113 QEYDPSKPNDYTAIHKEKMRQQQREELERLRQQELEKQKH 152
+EYDP++PNDY +EK R+ E++R E++K++
Sbjct 100 EEYDPARPNDYEEYKREKKRKATEAEMKR----EMDKRRQ 135
> dre:393889 rbm17, MGC55840, zgc:55840; RNA binding motif protein
17; K12840 splicing factor 45
Length=418
Score = 32.7 bits (73), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 16/33 (48%), Positives = 22/33 (66%), Gaps = 5/33 (15%)
Query 114 EYDPSKPNDYTAIHKEKMRQQQREELERLRQQE 146
EYDP PNDY EK+ ++ REE ++ R+QE
Sbjct 99 EYDPMFPNDY-----EKVLKRHREERQKQREQE 126
> dre:100331631 RNA binding motif protein 17-like
Length=418
Score = 32.3 bits (72), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 16/33 (48%), Positives = 22/33 (66%), Gaps = 5/33 (15%)
Query 114 EYDPSKPNDYTAIHKEKMRQQQREELERLRQQE 146
EYDP PNDY EK+ ++ REE ++ R+QE
Sbjct 99 EYDPMFPNDY-----EKVLKRHREERQKQREQE 126
> tgo:TGME49_043310 hypothetical protein
Length=1266
Score = 32.0 bits (71), Expect = 0.79, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 26/34 (76%), Gaps = 1/34 (2%)
Query 124 TAIHKEKMRQ-QQREELERLRQQELEKQKHDEEA 156
TA+ E+ ++ +QREE ER+R Q LE+++ +EEA
Sbjct 941 TALTAEETQELRQREEAERVRHQMLERKRKEEEA 974
> dre:564608 tmem198b, MGC112212, tmem198, zgc:112212; transmembrane
protein 198b
Length=369
Score = 30.8 bits (68), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 31/64 (48%), Gaps = 11/64 (17%)
Query 92 VTVGSSSPLDPTTWVPVGLWP---------QEYDPSKPNDYTAIHKEKMRQQQREELERL 142
V G P+ TTWV +G WP Q ++ +T + RQQ+R +L R+
Sbjct 208 VKTGPREPVCWTTWVVLGAWPALALLGVLVQWRVTAEGYSHTKVMIS--RQQRRVQLMRI 265
Query 143 RQQE 146
RQ+E
Sbjct 266 RQKE 269
> ath:AT4G21150 HAP6; HAP6 (HAPLESS 6); dolichyl-diphosphooligosaccharide-protein
glycotransferase; K12667 oligosaccharyltransferase
complex subunit delta (ribophorin II)
Length=691
Score = 30.8 bits (68), Expect = 1.7, Method: Composition-based stats.
Identities = 33/110 (30%), Positives = 45/110 (40%), Gaps = 8/110 (7%)
Query 9 MAKAAPKSSPSLNVQAAAADAALVLERLKKLHAGVLAEEQKPDGAASRAVASAAGSSAKA 68
+ +A PK S +QA DA L+L+ + VLA+EQ P S A A S K+
Sbjct 107 IGEAGPKDIVS-QLQAGVKDAKLLLDFYYSVRGLVLAKEQFPGTHISLGDAEAIFRSIKS 165
Query 69 GDVGAGGAESGHNETEPIVNVVSVTVGSSSPLDPTTWVPVGLWPQEYDPS 118
G N E S T + + V + L P E+DPS
Sbjct 166 LSQSDGRWRYSSNNPE------SSTFAAGLAYETLAGV-ISLAPSEFDPS 208
> mmu:244653 Hydin, 1700034M11Rik, 4930545D19Rik, 4932703P14,
A830061H17, AC069308.21gm4, hy-3, hy3, hyrh; hydrocephalus inducing
Length=5154
Score = 30.8 bits (68), Expect = 2.0, Method: Composition-based stats.
Identities = 18/53 (33%), Positives = 29/53 (54%), Gaps = 11/53 (20%)
Query 114 EYDPSKPNDYTAIHK---EKMRQQQREELERLR--------QQELEKQKHDEE 155
EYD + A + + +R++++ ELERL QQELE+QK ++E
Sbjct 2344 EYDALTAEEKVAFDRDVRQALRERKKRELERLAKEMQEKKLQQELERQKEEDE 2396
> cel:F58B3.7 hypothetical protein; K12840 splicing factor 45
Length=371
Score = 30.4 bits (67), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 26/42 (61%), Gaps = 1/42 (2%)
Query 114 EYDPSKPNDYTAIHKEKMRQQQREELERLRQQELEKQKHDEE 155
EY P PN+Y + KE ++QRE+ R + L+++ H+EE
Sbjct 113 EYYPMTPNNYEVVAKEINDRKQREKTAREVAKRLQRE-HEEE 153
> cel:T07C4.3 hypothetical protein
Length=649
Score = 29.6 bits (65), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 20/79 (25%), Positives = 37/79 (46%), Gaps = 1/79 (1%)
Query 22 VQAAAADAALVLERLKKLHAGVLAEEQKPDGAASRAVASAAGSSAKAGDVGAGGAESGHN 81
V A A+ + ++K AGV E KP ++ + GS+ + +V + + H
Sbjct 332 VSAELANGKPIKHLIEKFDAGVNFAEHKPKSIYAQVLEEIGGSAPRVDEVFSASKKEEHA 391
Query 82 ETEPIVNVVSVTVGSSSPL 100
ETE + NV+ + + S +
Sbjct 392 ETE-VTNVIFRSTSTHSSI 409
> hsa:94121 SYTL4, DKFZp451P0116, FLJ40960, SLP4; synaptotagmin-like
4
Length=671
Score = 28.9 bits (63), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 26/58 (44%), Gaps = 0/58 (0%)
Query 15 KSSPSLNVQAAAADAALVLERLKKLHAGVLAEEQKPDGAASRAVASAAGSSAKAGDVG 72
KS P LNV + ++ L KLH LA G++ + S ++AGD G
Sbjct 295 KSVPGLNVDMEEEEEEEDIDHLVKLHRQKLARSSMQSGSSMSTIGSMMSIYSEAGDFG 352
> mmu:22138 Ttn, 1100001C23Rik, 2310036G12Rik, 2310057K23Rik,
2310074I15Rik, AF006999, AV006427, D330041I19Rik, D830007G01Rik,
L56, mdm, shru; titin (EC:2.7.11.1); K12567 titin [EC:2.7.11.1]
Length=33467
Score = 28.9 bits (63), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 129 EKMRQQQREELERLRQQELEKQKHDEE 155
E ++Q Q EE RL +ELEK + DE+
Sbjct 8605 ELLKQSQEEETHRLEIEELEKSERDEK 8631
> hsa:50618 ITSN2, KIAA1256, PRO2015, SH3D1B, SH3P18, SWA, SWAP;
intersectin 2
Length=1697
Score = 28.5 bits (62), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 13/28 (46%), Positives = 22/28 (78%), Gaps = 0/28 (0%)
Query 128 KEKMRQQQREELERLRQQELEKQKHDEE 155
K+++ +Q+R E ER+R+QEL QK+ E+
Sbjct 451 KQELERQRRLEWERIRRQELLNQKNREQ 478
Lambda K H
0.305 0.122 0.335
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3451799900
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40