bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0999_orf3
Length=366
Score E
Sequences producing significant alignments: (Bits) Value
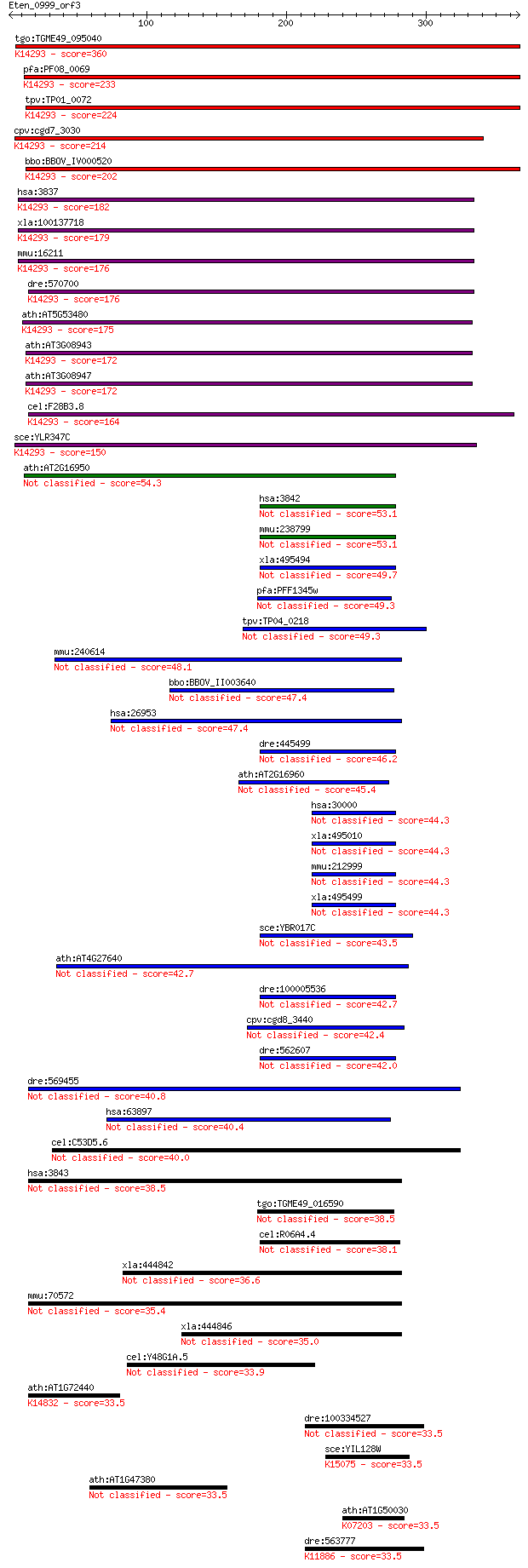
tgo:TGME49_095040 importin subunit beta-1, putative ; K14293 i... 360 6e-99
pfa:PF08_0069 importin beta, putative; K14293 importin subunit... 233 1e-60
tpv:TP01_0072 importin beta; K14293 importin subunit beta-1 224 6e-58
cpv:cgd7_3030 importin/karyopherin ; K14293 importin subunit b... 214 4e-55
bbo:BBOV_IV000520 21.m02834; importin beta subunit; K14293 imp... 202 2e-51
hsa:3837 KPNB1, IMB1, IPO1, IPOB, Impnb, MGC2155, MGC2156, MGC... 182 2e-45
xla:100137718 kpnb1, imb1, impnb, ipo1, ipob, ntf97; karyopher... 179 1e-44
mmu:16211 Kpnb1, AA409963, IPOB, Impnb, MGC8315; karyopherin (... 176 1e-43
dre:570700 kpnb1, MGC123184, wu:fc12b10, zgc:123184; karyopher... 176 1e-43
ath:AT5G53480 importin beta-2, putative; K14293 importin subun... 175 2e-43
ath:AT3G08943 binding / protein transporter; K14293 importin s... 172 2e-42
ath:AT3G08947 binding / protein transporter; K14293 importin s... 172 2e-42
cel:F28B3.8 imb-1; IMportin Beta family member (imb-1); K14293... 164 3e-40
sce:YLR347C KAP95, RSL1; Kap95p; K14293 importin subunit beta-1 150 1e-35
ath:AT2G16950 TRN1; TRN1 (TRANSPORTIN 1); protein transporter 54.3 7e-07
hsa:3842 TNPO1, IPO2, KPNB2, MIP, MIP1, TRN; transportin 1 53.1
mmu:238799 Tnpo1, AU021749, D13Ertd688e, IPO2, Kpnb2, MIP, MIP... 53.1 2e-06
xla:495494 tnpo1, ipo2, kpnb2, mip, mip1, trn; transportin 1 49.7
pfa:PFF1345w transportin 49.3 2e-05
tpv:TP04_0218 transportin 49.3 2e-05
mmu:240614 Ranbp6, C630001B19, FLJ11120; RAN binding protein 6 48.1
bbo:BBOV_II003640 18.m06305; transportin 47.4 8e-05
hsa:26953 RANBP6, FLJ54311; RAN binding protein 6 47.4
dre:445499 tnpo2, ik:tdsubc_2a7, tdsubc_2a7, wu:fb01c02, wu:fe... 46.2 2e-04
ath:AT2G16960 importin beta-2 subunit family protein 45.4 4e-04
hsa:30000 TNPO2, FLJ12155, IPO3, KPNB2B, TRN2; transportin 2 44.3
xla:495010 hypothetical LOC495010 44.3 8e-04
mmu:212999 Tnpo2, 1110034O24Rik, AA414969, AI464345, AI852433,... 44.3 8e-04
xla:495499 tnpo2; transportin 2 44.3
sce:YBR017C KAP104; Kap104p 43.5 0.001
ath:AT4G27640 importin beta-2 subunit family protein 42.7 0.002
dre:100005536 transportin-like 42.7 0.002
cpv:cgd8_3440 importin beta like ARM repeat alpha superhelix 42.4 0.003
dre:562607 tnpo1; transportin 1 42.0
dre:569455 kpnb3, cb273, wu:fc38a10; karyopherin (importin) be... 40.8 0.009
hsa:63897 HEATR6, ABC1, DKFZp686D22141, FLJ22087, MGC148096; H... 40.4 0.010
cel:C53D5.6 imb-3; IMportin Beta family member (imb-3) 40.0 0.014
hsa:3843 IPO5, DKFZp686O1576, FLJ43041, IMB3, KPNB3, MGC2068, ... 38.5 0.036
tgo:TGME49_016590 transportin, putative 38.5 0.037
cel:R06A4.4 imb-2; IMportin Beta family member (imb-2) 38.1 0.060
xla:444842 ipo5-a, MGC132286, ipo5, kap_beta_3, ranbp5; import... 36.6 0.16
mmu:70572 Ipo5, 1110011C18Rik, 5730478E03Rik, AA409333, C76941... 35.4 0.38
xla:444846 ipo5-b, ranbp5, ranbp6; importin 5 35.0
cel:Y48G1A.5 xpo-2; eXPOrtin (nuclear export receptor) family ... 33.9 1.1
ath:AT1G72440 EDA25; EDA25 (embryo sac development arrest 25);... 33.5 1.2
dre:100334527 si:dkey-245p14.6 33.5 1.2
sce:YIL128W MET18, MMS19; DNA repair and TFIIH regulator, requ... 33.5 1.3
ath:AT1G47380 protein phosphatase 2C-related / PP2C-related 33.5 1.4
ath:AT1G50030 TOR; TOR (TARGET OF RAPAMYCIN); 1-phosphatidylin... 33.5 1.4
dre:563777 similar to Proteasome-associated protein ECM29 homo... 33.5 1.4
> tgo:TGME49_095040 importin subunit beta-1, putative ; K14293
importin subunit beta-1
Length=971
Score = 360 bits (923), Expect = 6e-99, Method: Compositional matrix adjust.
Identities = 196/374 (52%), Positives = 249/374 (66%), Gaps = 30/374 (8%)
Query 6 GEDPSEALSQQRCNVVLTAVAHGLKDPDPQLKVAALRALYHALIFAKENFAKTEERDYII 65
GEDP++ LS+ CN +LTAV G+KD D QLKVAAL+ALYHALIF+K+NF ER+YII
Sbjct 207 GEDPADVLSEAHCNNILTAVVQGMKDEDVQLKVAALKALYHALIFSKKNFENQTEREYII 266
Query 66 SSVLEAGNAANPSAVQVAALECIVQISQEYYSFLGAYMPAVGSVTWNALKSSDSAVVIAA 125
VLE A+ AVQV+A EC+V++++EYYS L YM VG ++W ALKS D++V IAA
Sbjct 267 QVVLENTKVAH-QAVQVSAFECLVKVAEEYYSMLEPYMSGVGPLSWEALKSGDASVCIAA 325
Query 126 LEVWNALAEEELAIIEGESTRWTL------------LNIMKEATPFLLPLLLEQLLNA-A 172
+E+WN +A+ E+ I + E L +I+K+A PFLLP+LL L +
Sbjct 326 MELWNTIADVEIDIQQQEEEAAALGADGAESAVPRNCHIVKQALPFLLPILLNTLTQQDS 385
Query 173 EDEDDDDSWTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCV 232
E+ D DSWT AMAA CLGLCAQVVK+ ILP VI FV NF S +W RREA+VLA G V
Sbjct 386 EETDAADSWTAAMAAGTCLGLCAQVVKNDILPPVIQFVSENFASPDWTRREAAVLAFGSV 445
Query 233 MEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPAD 292
MEGP EA+ P V SF LV+ V D+SVAVRD+AAWTLG+ AQ H P+VL+ L AD
Sbjct 446 MEGPDTEALKPLVEESFASLVD-VLQDSSVAVRDTAAWTLGRIAQFHTPVVLQKLV-NAD 503
Query 293 VSSTTSPNNNGGALLLAIVERLTDQPRVAVHICWMLHELADDINTPEGRAAYQGEAATQQ 352
+ N +LL AIV RL DQPRVAV++CW+LHELAD + +G ++
Sbjct 504 GTVVVESN----SLLAAIVRRLLDQPRVAVNVCWLLHELADHMTAGDG----------ER 549
Query 353 QDQTPLDPIFPKLC 366
TPLDP+F +LC
Sbjct 550 PASTPLDPLFQRLC 563
> pfa:PF08_0069 importin beta, putative; K14293 importin subunit
beta-1
Length=877
Score = 233 bits (593), Expect = 1e-60, Method: Compositional matrix adjust.
Identities = 133/361 (36%), Positives = 202/361 (55%), Gaps = 29/361 (8%)
Query 12 ALSQQRCNVVLTAVAHGLKDP-DPQLKVAALRALYHALIFAKENFAKTEERDYIISSVLE 70
A +Q +++LTA+ + L +P + A ++ LY+ + F + NF ERD I+ +V++
Sbjct 176 AFTQPDLDLILTAIINSLCEPAEESTHCANMKVLYNLMSFIEHNFKTQVERDIIMKTVID 235
Query 71 AGNAANPSAVQVAALECIVQISQEYYSFLGAYMPAVGSVTWNALKSSDSAVVIAALEVWN 130
+VQ+AA EC++ I +YS+L AYM A+G +TW A++S + + I+A+E WN
Sbjct 236 GCKDTERQSVQIAAYECLINIVSYFYSYLDAYMYAIGPLTWVAIESENERIAISAIEFWN 295
Query 131 ALAEEELAI-----IEGESTRWTLLNIMKEATPFLLPLLLEQLLNAAEDEDDDDSWTPAM 185
+ EEE I EG+ NI+K+A FLLP + ++ ++ D D+WT +M
Sbjct 296 TVCEEETFIDQYELQEGKKNH----NIVKQAMVFLLPKIFNAMITQESEDIDIDAWTLSM 351
Query 186 AAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEAMAPYV 245
A+A L L AQ++K+ I+ VI+FV+ NF +WRRR+A+VLA G +MEGP E + P V
Sbjct 352 ASATFLALSAQLLKNDIVEPVISFVEENFIHEDWRRRDAAVLAYGSIMEGPDTEKLKPLV 411
Query 246 ARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPADVSSTTSPNNNGGA 305
S L E V D SVAVRD+AAWT+GK H+ I+ L N+ +
Sbjct 412 EESVGQLSE-VLRDPSVAVRDTAAWTIGKITTYHSEIIYNVL----------GSYNDSNS 460
Query 306 LLLAIVERLTDQPRVAVHICWMLHELADDINTPEGRAAYQGEAATQQQDQTPLDPIFPKL 365
L ++ERL D PRVA ++CW+ ++LA R +Y + D LD F L
Sbjct 461 LYGILLERLNDYPRVAANVCWVFNQLA-----VNKRTSYNKMTNSYVTD---LDDSFCVL 512
Query 366 C 366
C
Sbjct 513 C 513
> tpv:TP01_0072 importin beta; K14293 importin subunit beta-1
Length=873
Score = 224 bits (570), Expect = 6e-58, Method: Compositional matrix adjust.
Identities = 143/371 (38%), Positives = 199/371 (53%), Gaps = 41/371 (11%)
Query 13 LSQQRCNVVLTAVAHGLKDPDPQLKVAALRALYHALIFAKENFAKTEERDYIISSVLEAG 72
LS N +LT+V G+ DPQ ALR+L + L F + N ERD I+ ++
Sbjct 172 LSDLEVNRLLTSVIKGVYIEDPQSCKMALRSLQNLLFFIENNMEVDAERDVIVEAICRRC 231
Query 73 NAANPSAVQVAALECIVQISQEYYSFLGAYMPAVGSVTWNALKSSDSAVVIAALEVWNAL 132
+ N ++ AA +C+VQ+ EYYS L + + W A+ S + I A E WN +
Sbjct 232 SENNDLEIRTAAFDCLVQLVSEYYSRLIPSLQVIVPFLWQAIDSHVEQIAIPAFEFWNTI 291
Query 133 AEEEL-----------AIIEGESTRWTLLN-----IMKEATPFLLP-LLLEQLLNAAEDE 175
E E+ + + EST + + I+K+ P+LLP +L L+ ED
Sbjct 292 CEIEIQSAANATDRTSSTVRSESTGKSNRDAVEGSIIKQVIPYLLPKILFTMTLHKFEDM 351
Query 176 DDDDSWTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEG 235
D D +WT MAA ICL LC+Q VK+ I+ V+ FV NF STEW +REA+VLA G +MEG
Sbjct 352 DVD-TWTLPMAAGICLSLCSQTVKNDIVHSVLEFVTENFKSTEWNKREAAVLAYGYIMEG 410
Query 236 PSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPADVSS 295
P +E + V+ SF L + V SD S+AVRD+AAWT+G+ A H +VL HL P DV
Sbjct 411 PDSETLKILVSESFDNLCD-VLSDTSIAVRDTAAWTIGRIATFHCEVVLNHLGSP-DV-- 466
Query 296 TTSPNNNGGALLLAIVERLTDQPRVAVHICWMLHELADDINTPEGRAAYQGEAATQQQDQ 355
PN+N L IV L D PRVAV+IC+ ++ELA+ IN +
Sbjct 467 ---PNSN----LSKIVRALFDVPRVAVNICYFINELAEHIND------------YNKGPT 507
Query 356 TPLDPIFPKLC 366
LD +F +LC
Sbjct 508 NLLDCMFARLC 518
> cpv:cgd7_3030 importin/karyopherin ; K14293 importin subunit
beta-1
Length=882
Score = 214 bits (545), Expect = 4e-55, Method: Compositional matrix adjust.
Identities = 123/343 (35%), Positives = 192/343 (55%), Gaps = 13/343 (3%)
Query 5 EGEDPSEALSQQRCNVVLTAVAHGLKDPDPQLKVAALRALYHALIFAKENFAKTEERDYI 64
E E S +++ N +LTA+ G+ DPD + +AA ++ Y+AL FA+ NF+ ER+ I
Sbjct 169 ENEVSSLIITEDISNQILTAIVQGMNDPDSETALAATKSFYYALYFARSNFSNEMERNLI 228
Query 65 ISSVLE-AGNAANPSAV-QVAALECIVQISQEYYSFLGAYMPAVGSVTWNALKSSDSAVV 122
+ G N + Q AA EC+V I+ EYY +LG+Y+ + +T +K V
Sbjct 229 FQVLCTLCGTEGNKRELLQTAAYECLVSIATEYYDYLGSYLSVLTPMTIKGIKGVYEPVS 288
Query 123 IAALEVWNALAEEELAI-IEGE----STRWTLLNIMKEATPFLLPLLLEQLLNAAEDEDD 177
I +E WN +A+ E+ + +E E S + ++ + + L+P++LE LL D+DD
Sbjct 289 ICCIEFWNTIADLEIELSLEDEHNNTSPSTSCMHYISQVQAALIPVMLETLLRQ-NDDDD 347
Query 178 DDSWTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPS 237
+SWT + AA CL LC+Q++ D IL + F+ NF + W REA+VLA G ++EGPS
Sbjct 348 LESWTVSKAAGACLTLCSQLLGDNILEPTLGFIHSNFSHSNWHNREAAVLAYGSILEGPS 407
Query 238 AEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPADVSSTT 297
+ M P V S L +A+ +D VAVRD+ AWT+G+ H I+ FP +
Sbjct 408 IQKMQPIVETSVTNLCQAL-NDNVVAVRDTCAWTIGRIVTFHPTII----FPLLGLPPQQ 462
Query 298 SPNNNGGALLLAIVERLTDQPRVAVHICWMLHELADDINTPEG 340
S ++ G L L + L D+PRV +ICW++H++A+ EG
Sbjct 463 SVEHSNGLLSLLLQRLLADEPRVCTNICWIIHQIAESSQYVEG 505
> bbo:BBOV_IV000520 21.m02834; importin beta subunit; K14293 importin
subunit beta-1
Length=859
Score = 202 bits (514), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 119/356 (33%), Positives = 185/356 (51%), Gaps = 30/356 (8%)
Query 13 LSQQRCNVVLTAVAHGLKDPDPQLKVAALRALYHALIFAKENFAKTEERDYIISSVLEAG 72
LS+ + +LT++ G D Q + + + H L F N A ERD I+ ++
Sbjct 174 LSKAESDRLLTSIVKGAFMSDTQSRRTVMICMQHILPFVDSNMAIPNERDTIVQAICYNS 233
Query 73 NAANPSAVQVAALECIVQISQEYYSFLGAYMPAVGSVTWNALKSSDSAVVIAALEVWNAL 132
+NPS ++ AA +C+VQ+ +YY +G + + + W + + V I A E WN +
Sbjct 234 GPSNPSNLRAAANDCLVQLVTDYYEVIGPCLQYIVPLLWEGIDTRIEEVAIPAFEFWNTI 293
Query 133 AEEELAII--EGESTRWTLLNIMKEATPFLLPLLLEQLLNAAEDEDDDDSWTPAMAAAIC 190
E E+A+ ES + +I+++ +LLP +L + ++ D D+WT MAA +C
Sbjct 294 CETEIALSYENSESNQ----HIIQQVISYLLPKILFTMTLHEFEDFDSDTWTLPMAAGVC 349
Query 191 LGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEAMAPYVARSFP 250
L LC+Q VK+ I+P V+ F+ NF +W REA+VLA G +MEGP A+ + V SF
Sbjct 350 LSLCSQAVKNDIVPSVLQFINENFHHAQWNCREAAVLAYGYIMEGPDADTLRLLVRDSFD 409
Query 251 ILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPADVSSTTSPNNNGGALLLAI 310
L + V D S+AV+D+AAWT+G+ A H ++L HL S P +N + I
Sbjct 410 RLCD-VLDDPSIAVQDTAAWTIGRIASFHCAVILPHL------GSLEDPGSN----ICKI 458
Query 311 VERLTDQPRVAVHICWMLHELADDINTPEGRAAYQGEAATQQQDQTPLDPIFPKLC 366
+ L RVA +ICW HELA+ + E + LD +FPK+C
Sbjct 459 MRALFKPARVAANICWFFHELAEGLGQIEA-------------SRYMLDAMFPKIC 501
> hsa:3837 KPNB1, IMB1, IPO1, IPOB, Impnb, MGC2155, MGC2156, MGC2157,
NTF97; karyopherin (importin) beta 1; K14293 importin
subunit beta-1
Length=876
Score = 182 bits (463), Expect = 2e-45, Method: Compositional matrix adjust.
Identities = 117/337 (34%), Positives = 186/337 (55%), Gaps = 30/337 (8%)
Query 8 DPSEALSQQRCNVVLTAVAHGLKDPDP--QLKVAALRALYHALIFAKENFAKTEERDYII 65
DP + Q + N +LTA+ G++ +P +K+AA AL ++L F K NF K ER +I+
Sbjct 162 DPEQL--QDKSNEILTAIIQGMRKEEPSNNVKLAATNALLNSLEFTKANFDKESERHFIM 219
Query 66 SSVLEAGNAANPSAVQVAALECIVQISQEYYSFLGAYM-PAVGSVTWNALKSSDSAVVIA 124
V EA + + V+VAAL+ +V+I YY ++ YM PA+ ++T A+KS V +
Sbjct 220 QVVCEATQCPD-TRVRVAALQNLVKIMSLYYQYMETYMGPALFAITIEAMKSDIDEVALQ 278
Query 125 ALEVWNALAEEEL--------AIIEGESTRWTLLNIMKEATPFLLPLLLEQLLNAAEDED 176
+E W+ + +EE+ A +G T K A +L+P+L Q L ++ D
Sbjct 279 GIEFWSNVCDEEMDLAIEASEAAEQGRPPEHTSKFYAKGALQYLVPIL-TQTLTKQDEND 337
Query 177 DDDSWTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGP 236
DDD W P AA +CL L A +D I+P V+ F++ + + +WR R+A+V+A GC++EGP
Sbjct 338 DDDDWNPCKAAGVCLMLLATCCEDDIVPHVLPFIKEHIKNPDWRYRDAAVMAFGCILEGP 397
Query 237 SAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPADVSST 296
+ P V ++ P L+E + D SV VRD+AAWT+G+ + L P A ++
Sbjct 398 EPSQLKPLVIQAMPTLIE-LMKDPSVVVRDTAAWTVGRICE---------LLPEAAINDV 447
Query 297 TSPNNNGGALLLAIVERLTDQPRVAVHICWMLHELAD 333
LL ++E L+ +PRVA ++CW LA+
Sbjct 448 YL-----APLLQCLIEGLSAEPRVASNVCWAFSSLAE 479
> xla:100137718 kpnb1, imb1, impnb, ipo1, ipob, ntf97; karyopherin
(importin) beta 1; K14293 importin subunit beta-1
Length=876
Score = 179 bits (455), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 115/337 (34%), Positives = 185/337 (54%), Gaps = 30/337 (8%)
Query 8 DPSEALSQQRCNVVLTAVAHGLKDPDP--QLKVAALRALYHALIFAKENFAKTEERDYII 65
DP + Q + N +LTA+ G++ +P +++AA AL ++L F K NF K ER YI+
Sbjct 162 DPEQL--QHKSNEILTAIIQGMRKEEPSNNVRLAATNALLNSLEFTKANFDKESERHYIM 219
Query 66 SSVLEAGNAANPSAVQVAALECIVQISQEYYSFLGAYM-PAVGSVTWNALKSSDSAVVIA 124
V EA + + V+VAAL+ +V+I YY ++ YM PA+ ++T A+K+ V +
Sbjct 220 QVVCEATQCPD-TRVRVAALQNLVKIMSLYYQYMETYMGPALFAITVEAMKNEIDEVALQ 278
Query 125 ALEVWNALAEEEL--------AIIEGESTRWTLLNIMKEATPFLLPLLLEQLLNAAEDED 176
+E W+ + +EE+ A +G T K A +L+P+L Q L ++ D
Sbjct 279 GIEFWSNVCDEEMDLAIEASEAAEQGRPPEHTSKFYAKGALQYLVPIL-TQTLTKQDEND 337
Query 177 DDDSWTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGP 236
DDD W P AA +CL L A +D I+P V+ F++ + + +WR R+A+V+A GC++EGP
Sbjct 338 DDDDWNPCKAAGVCLMLLATCCEDDIVPHVLPFIKEHIKNPDWRYRDAAVMAFGCILEGP 397
Query 237 SAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPADVSST 296
+ + P V ++ P L+E + D SV VRD+ AWT+G+ + L P A ++
Sbjct 398 ESCQLKPLVIQAMPTLIE-LMKDPSVVVRDTTAWTVGRICE---------LLPEAAINDV 447
Query 297 TSPNNNGGALLLAIVERLTDQPRVAVHICWMLHELAD 333
LL ++E L +PRVA ++CW LA+
Sbjct 448 YL-----APLLQCLIEGLGAEPRVASNVCWAFSSLAE 479
> mmu:16211 Kpnb1, AA409963, IPOB, Impnb, MGC8315; karyopherin
(importin) beta 1; K14293 importin subunit beta-1
Length=876
Score = 176 bits (446), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 114/337 (33%), Positives = 184/337 (54%), Gaps = 30/337 (8%)
Query 8 DPSEALSQQRCNVVLTAVAHGLKDPDP--QLKVAALRALYHALIFAKENFAKTEERDYII 65
DP + Q + N +LTA+ G++ +P +K+AA AL ++L F K NF K ER +I+
Sbjct 162 DPEQL--QDKSNEILTAIIQGMRKEEPSNNVKLAATNALLNSLEFTKANFDKESERHFIM 219
Query 66 SSVLEAGNAANPSAVQVAALECIVQISQEYYSFLGAYM-PAVGSVTWNALKSSDSAVVIA 124
V EA + + V+VAAL+ +V+I YY ++ YM PA+ ++T A+KS V +
Sbjct 220 QVVCEATQCPD-TRVRVAALQNLVKIMSLYYQYMETYMGPALFAITIEAMKSDIDEVALQ 278
Query 125 ALEVWNALAEEEL--------AIIEGESTRWTLLNIMKEATPFLLPLLLEQLLNAAEDED 176
+E W+ + +EE+ A +G T K A +L+P+L Q L ++ D
Sbjct 279 GIEFWSNVCDEEMDLAIEASEAAEQGRPPEHTSKFYAKGALQYLVPIL-TQTLTKQDEND 337
Query 177 DDDSWTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGP 236
DDD W P AA +CL L + +D I+P V+ F++ + + +WR R+A+V+A G ++EGP
Sbjct 338 DDDDWNPCKAAGVCLMLLSTCCEDDIVPHVLPFIKEHIKNPDWRYRDAAVMAFGSILEGP 397
Query 237 SAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPADVSST 296
+ P V ++ P L+E + D SV VRD+ AWT+G+ + L P A ++
Sbjct 398 EPNQLKPLVIQAMPTLIE-LMKDPSVVVRDTTAWTVGRICE---------LLPEAAINDV 447
Query 297 TSPNNNGGALLLAIVERLTDQPRVAVHICWMLHELAD 333
LL ++E L+ +PRVA ++CW LA+
Sbjct 448 YL-----APLLQCLIEGLSAEPRVASNVCWAFSSLAE 479
> dre:570700 kpnb1, MGC123184, wu:fc12b10, zgc:123184; karyopherin
(importin) beta 1; K14293 importin subunit beta-1
Length=876
Score = 176 bits (446), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 115/331 (34%), Positives = 179/331 (54%), Gaps = 30/331 (9%)
Query 15 QQRCNVVLTAVAHGLKDPDP--QLKVAALRALYHALIFAKENFAKTEERDYIISSVLEAG 72
Q N +LTA+ G++ +P +K+AA AL ++L F K NF K ER +I+ V EA
Sbjct 167 QDSANQILTAIIQGMRKEEPSNNVKLAATNALLNSLEFTKANFDKETERHFIMQVVCEAT 226
Query 73 NAANPSAVQVAALECIVQISQEYYSFLGAYM-PAVGSVTWNALKSSDSAVVIAALEVWNA 131
+ + V+VAAL+ +V+I YY ++ YM PA+ ++T A+KS V + +E W+
Sbjct 227 QCPD-TRVRVAALQNLVKIMSLYYQYMETYMGPALFAITIEAMKSDIDEVALQGIEFWSN 285
Query 132 LAEEELAII--------EGESTRWTLLNIMKEATPFLLPLLLEQLLNAAEDEDDDDSWTP 183
+ +EE+ + +G T K A +L+P+L Q L ++ DDDD W P
Sbjct 286 VCDEEMDLAIEATEASEQGRPPEHTSKFYAKGALQYLVPIL-TQTLTKQDENDDDDDWNP 344
Query 184 AMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEAMAP 243
AA +CL L A +D ++P V+ F++ + +WR R+ASV+A G ++EGP + P
Sbjct 345 CKAAGVCLMLLATCCEDDVVPHVLPFIKEHIKHPDWRYRDASVMAFGSILEGPELNQLKP 404
Query 244 YVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPADVSST-TSPNNN 302
V ++ P L+E + D SV VRD+ AWTLG+ L P A ++ SP
Sbjct 405 LVIQAMPTLIE-LMKDPSVVVRDTTAWTLGRICD---------LLPEAAINEVYLSP--- 451
Query 303 GGALLLAIVERLTDQPRVAVHICWMLHELAD 333
LL ++E L +PRVA ++CW LA+
Sbjct 452 ---LLQCLIEGLGAEPRVASNVCWAFSSLAE 479
> ath:AT5G53480 importin beta-2, putative; K14293 importin subunit
beta-1
Length=870
Score = 175 bits (444), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 114/334 (34%), Positives = 180/334 (53%), Gaps = 25/334 (7%)
Query 11 EALSQQRCNVVLTAVAHGLK--DPDPQLKVAALRALYHALIFAKENFAKTEERDYIISSV 68
+ + Q+ N +LTAV G+ + + +++AA RALY AL FA+ NF ERDYI+ V
Sbjct 165 DVVEQEHVNKILTAVVQGMNAAEGNTDVRLAATRALYMALGFAQANFNNDMERDYIMRVV 224
Query 69 LEAGNAANPSAVQVAALECIVQISQEYYSFLGAYMPAVGSVTWNALKSSDSAVVIAALEV 128
EA + ++ AA EC+V I+ YY L YM + ++T A++ D +V + A+E
Sbjct 225 CEA-TLSPEVKIRQAAFECLVSIASTYYEKLAHYMQDIFNITAKAVREDDESVALQAIEF 283
Query 129 WNALAEEELAIIEGESTRWT------LLNIMKEATPFLLPLLLEQLLNAAEDED-DDDSW 181
W+++ +EE+ I+E + K+A P L+PLLLE LL ED+D D+ +W
Sbjct 284 WSSICDEEIDILEEYGGEFAGDSDVPCFYFTKQALPGLVPLLLETLLKQEEDQDLDEGAW 343
Query 182 TPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEAM 241
AMA CLGL A+ V D I+P V+ F++ +WR REA+ A G ++EGPSA+ +
Sbjct 344 NIAMAGGTCLGLVARAVGDDIVPHVMPFIEEKISKPDWREREAATYAFGSILEGPSADKL 403
Query 242 APYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPADVSSTTSP-- 299
V + ++ A+ +D S V+D+ AWTLG+ +F S+ +P
Sbjct 404 MAIVNAALTFMLNALTNDPSNHVKDTTAWTLGR------------IFEFLHGSTIETPII 451
Query 300 -NNNGGALLLAIVERLTDQPRVAVHICWMLHELA 332
N ++ +++ + D P VA C L+ LA
Sbjct 452 NQANCQQIITVLIQSMNDAPNVAEKACGALYFLA 485
> ath:AT3G08943 binding / protein transporter; K14293 importin
subunit beta-1
Length=871
Score = 172 bits (436), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 117/329 (35%), Positives = 179/329 (54%), Gaps = 21/329 (6%)
Query 13 LSQQRCNVVLTAVAHGLKDPD--PQLKVAALRALYHALIFAKENFAKTEERDYIISSVLE 70
L Q N VLTAV G+ + ++++AA +AL +AL F++ NF ER+YI+ V E
Sbjct 169 LVQDEVNSVLTAVVQGMNQSENTAEVRLAATKALLNALDFSQTNFENEMERNYIMKMVCE 228
Query 71 AGNAANPSAVQVAALECIVQISQEYYSFLGAYMPAVGSVTWNALKSSDSAVVIAALEVWN 130
+ + ++ AA EC+V I+ YY L Y+ + +T NA+K + +V + A+E W+
Sbjct 229 TA-CSKEAEIRQAAFECLVSIASTYYEVLEHYIQTLFELTSNAVKGDEESVALQAIEFWS 287
Query 131 ALAEEELAIIE------GESTRWTLLNIMKEATPFLLPLLLEQLLNAAEDEDDDDS-WTP 183
++ +EE+ E G+S+ I K A P L+ +LLE LL ED+D DD W
Sbjct 288 SICDEEIDRQEYDSPDSGDSSPPHSCFIEK-ALPHLVQMLLETLLKQEEDQDHDDDVWNI 346
Query 184 AMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEAMAP 243
+MA CLGL A+ V D ++PLV+ FV+ N S +WR REA+ A G ++EGP+ + +AP
Sbjct 347 SMAGGTCLGLVARTVGDGVVPLVMPFVEKNISSPDWRSREAATYAFGSILEGPTIDKLAP 406
Query 244 YVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPADVSSTTSPNNNG 303
VA L+ A D + VRD+ AWTL + + L P S SP N
Sbjct 407 MVAAGLEFLLNAT-KDQNNHVRDTTAWTLSR--------IFEFLPSPDSGFSVISP-ENL 456
Query 304 GALLLAIVERLTDQPRVAVHICWMLHELA 332
++ ++E + D P VA +C ++ LA
Sbjct 457 PRIVSVLLESIKDVPNVAEKVCGAIYNLA 485
> ath:AT3G08947 binding / protein transporter; K14293 importin
subunit beta-1
Length=873
Score = 172 bits (435), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 114/328 (34%), Positives = 180/328 (54%), Gaps = 19/328 (5%)
Query 13 LSQQRCNVVLTAVAHGLKDPD--PQLKVAALRALYHALIFAKENFAKTEERDYIISSVLE 70
L Q N VLTAV G+ + ++++AA +AL +AL F++ NF ER+YI+ V E
Sbjct 169 LVQDEVNSVLTAVVQGMNQSENTAEVRLAATKALCNALDFSQTNFENEMERNYIMKMVCE 228
Query 71 AGNAANPSAVQVAALECIVQISQEYYSFLGAYMPAVGSVTWNALKSSDSAVVIAALEVWN 130
+ + ++ AA EC+V I+ YY L Y+ + +T NA+K + +V + A+E W+
Sbjct 229 TA-CSKEAEIRQAAFECLVSIASTYYEVLEHYIQTLFELTSNAVKGDEESVSLQAIEFWS 287
Query 131 ALAEEELAIIEGES-----TRWTLLNIMKEATPFLLPLLLEQLLNAAEDEDDDDS-WTPA 184
++ +EE+ E +S + + +++A P L+ +LLE LL ED+D DD W +
Sbjct 288 SICDEEIDRQEYDSPASGDSSPPHSSFIEKALPHLVQMLLETLLKQEEDQDHDDDVWNIS 347
Query 185 MAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEAMAPY 244
MA CLGL A+ V D ++PLV+ FV+ N S +WR REA+ A G ++EGP+ + +AP
Sbjct 348 MAGGTCLGLVARTVGDHVVPLVMPFVEKNISSPDWRCREAATYAFGSILEGPTIDKLAPM 407
Query 245 VARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPADVSSTTSPNNNGG 304
VA L+ A D + VRD+ AWTL + + L P S SP N
Sbjct 408 VAAGLEFLLNAT-KDQNNHVRDTTAWTLSR--------IFEFLHSPDSGFSVISP-ENLP 457
Query 305 ALLLAIVERLTDQPRVAVHICWMLHELA 332
++ ++E + D P VA +C ++ LA
Sbjct 458 RIVSVLLESIKDVPNVAEKVCGAIYNLA 485
> cel:F28B3.8 imb-1; IMportin Beta family member (imb-1); K14293
importin subunit beta-1
Length=896
Score = 164 bits (416), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 118/364 (32%), Positives = 195/364 (53%), Gaps = 41/364 (11%)
Query 15 QQRCNVVLTAVAHGLK--DPDPQLKVAALRALYHALIFAKENFAKTEERDYIISSVLEAG 72
+ + N VLTA+ HG++ + ++ AA AL ++L F NF+ ER+ I+ V E+
Sbjct 173 ETKANDVLTAIIHGMRPEESSANVRFAATNALLNSLEFTNTNFSNEAERNIIMQVVCEST 232
Query 73 NAANPSAVQVAALECIVQISQEYYSFLGAYM-PAVGSVTWNALKSSDSAVVIAALEVWNA 131
++++ V+VAAL+C+V+I Q YY + +YM A+ +T +A+KS + V + +E W+
Sbjct 233 SSSD-QRVKVAALQCLVRIMQLYYEHMLSYMGSALFQITLSAMKSQEPEVAMQGMEFWST 291
Query 132 LAEEELAII---EGESTR------WTLLNIMKEATPFLLPLLLEQLLNAAEDEDDDDSWT 182
+AEEE + E E R L M++A + P+LLE + + + +DDD WT
Sbjct 292 VAEEEFDLYMTYEDEVERGAPNPKCASLRFMEQAASHVCPVLLEAMAHHDDGDDDD-DWT 350
Query 183 PAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEAMA 242
PA AA +CL L AQ V+D I+ VI F + +F + +W+ +EA+++A G +++GP + +
Sbjct 351 PAKAAGVCLMLAAQCVRDDIVNHVIPFFK-HFQNPDWKYKEAAIMAFGSILDGPDPKKLL 409
Query 243 PYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPADVSSTTSPNNN 302
P + P +V A+ D +V VRD+AAW+LG+ V T S N
Sbjct 410 PMAQEALPAIVAAMC-DKNVNVRDTAAWSLGR------------------VIDTCSELAN 450
Query 303 GGALLLAIVERLTD----QPRVAVHICWMLHELADDINTPEGRAAYQGEAATQQQDQTPL 358
LL +++ L++ +PRVA ++CW L L + A G + Q D L
Sbjct 451 NAELLQSVLPVLSNGLHQEPRVANNVCWALVSL---VKACYESAVANGTDGSGQPDTFAL 507
Query 359 DPIF 362
+F
Sbjct 508 SSVF 511
> sce:YLR347C KAP95, RSL1; Kap95p; K14293 importin subunit beta-1
Length=861
Score = 150 bits (378), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 106/343 (30%), Positives = 170/343 (49%), Gaps = 34/343 (9%)
Query 5 EGEDPSEALSQQRCNVVLTAVAHGLKDPDPQ--LKVAALRALYHALIFAKENFAKTEERD 62
E DP N +L A+ G + + +++AAL AL +LIF K N + ER+
Sbjct 164 ESADPQSQALVSSSNNILIAIVQGAQSTETSKAVRLAALNALADSLIFIKNNMEREGERN 223
Query 63 YIISSVLEAGNAANPSAVQVAALECIVQISQEYYSFLGAYMP-AVGSVTWNALKSSDSAV 121
Y++ V EA A + VQ AA C+ +I YY+F+ YM A+ ++T +KS + V
Sbjct 224 YLMQVVCEATQAEDIE-VQAAAFGCLCKIMSLYYTFMKPYMEQALYALTIATMKSPNDKV 282
Query 122 VIAALEVWNALAEEELAIIEG---------ESTRWTLLNIMKEATPFLLPLLLEQLLNAA 172
+E W+ + EEE+ I +S + L +I K+ P LL LL Q
Sbjct 283 ASMTVEFWSTICEEEIDIAYELAQFPQSPLQSYNFALSSI-KDVVPNLLNLLTRQ----N 337
Query 173 EDEDDDDSWTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCV 232
ED +DDD W +M+A CL L AQ + IL V+ FV+ N + WR REA+V+A G +
Sbjct 338 EDPEDDD-WNVSMSAGACLQLFAQNCGNHILEPVLEFVEQNITADNWRNREAAVMAFGSI 396
Query 233 MEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPAD 292
M+GP YV ++ P ++ + +D S+ V+++ AW +G+ A
Sbjct 397 MDGPDKVQRTYYVHQALPSILN-LMNDQSLQVKETTAWCIGRIAD--------------S 441
Query 293 VSSTTSPNNNGGALLLAIVERLTDQPRVAVHICWMLHELADDI 335
V+ + P + ++ A + L D P+VA + W + L + +
Sbjct 442 VAESIDPQQHLPGVVQACLIGLQDHPKVATNCSWTIINLVEQL 484
> ath:AT2G16950 TRN1; TRN1 (TRANSPORTIN 1); protein transporter
Length=891
Score = 54.3 bits (129), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 68/309 (22%), Positives = 118/309 (38%), Gaps = 61/309 (19%)
Query 12 ALSQQRCNVVLTAVAHGLKDPDPQLKVAALRALYHALIFAKENFAKTEERDYIISSVLEA 71
L+++ N+ L + + P L+ AL ++ +I + ++ VL
Sbjct 173 GLAERPINIFLPRLLQFFQSPHASLRKLALGSVNQYIIIMPAALYNSLDKYLQGLFVL-- 230
Query 72 GNAANPSAVQVAALEC--IVQISQEYYSFLGAYMPAVGSVTWNALKSSDSAVVIAALEVW 129
AN +V L C V +++ S + ++ V + D V + A E W
Sbjct 231 ---ANDPVPEVRKLVCAAFVHLTEVLPSSIEPHLRNVMEYMLQVNRDPDEEVSLEACEFW 287
Query 130 NALAEEELAIIEGESTRWTLLNIMKEATPFLLPLLLEQ---------LLNAAEDEDDDDS 180
+A + +L +KE P L+P+LLE LL+A EDE D
Sbjct 288 SAYCDAQLPP-----------ENLKEFLPRLIPVLLENMAYADDDESLLDAEEDESQPDR 336
Query 181 -----------------------------WTPAMAAAICLGLCAQVVKDAILPLVINFVQ 211
W +A + + + V D ILP ++ +Q
Sbjct 337 DQDLKPRFHTSRLHGSEDFDDDDDDSFNVWNLRKCSAAAIDVLSNVFGDEILPALMPLIQ 396
Query 212 LNF---GSTEWRRREASVLALGCVMEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSA 268
N G W++REA+VLALG + EG + P+++ L+ + D +R +
Sbjct 397 KNLSASGDEAWKQREAAVLALGAIAEG-CMNGLYPHLSEIVAFLL-PLLDDKFPLIRSIS 454
Query 269 AWTLGKTAQ 277
WTL + +
Sbjct 455 CWTLSRFGK 463
> hsa:3842 TNPO1, IPO2, KPNB2, MIP, MIP1, TRN; transportin 1
Length=898
Score = 53.1 bits (126), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/97 (29%), Positives = 47/97 (48%), Gaps = 2/97 (2%)
Query 181 WTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEA 240
W +A L + A V +D +LP ++ ++ EW +E+ +L LG + EG +
Sbjct 381 WNLRKCSAAALDVLANVYRDELLPHILPLLKELLFHHEWVVKESGILVLGAIAEG-CMQG 439
Query 241 MAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQ 277
M PY+ P L++ + SD VR WTL + A
Sbjct 440 MIPYLPELIPHLIQCL-SDKKALVRSITCWTLSRYAH 475
> mmu:238799 Tnpo1, AU021749, D13Ertd688e, IPO2, Kpnb2, MIP, MIP1,
TRN; transportin 1
Length=890
Score = 53.1 bits (126), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 29/97 (29%), Positives = 47/97 (48%), Gaps = 2/97 (2%)
Query 181 WTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEA 240
W +A L + A V +D +LP ++ ++ EW +E+ +L LG + EG +
Sbjct 373 WNLRKCSAAALDVLANVYRDELLPHILPLLKELLFHHEWVVKESGILVLGAIAEG-CMQG 431
Query 241 MAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQ 277
M PY+ P L++ + SD VR WTL + A
Sbjct 432 MIPYLPELIPHLIQCL-SDKKALVRSITCWTLSRYAH 467
> xla:495494 tnpo1, ipo2, kpnb2, mip, mip1, trn; transportin 1
Length=890
Score = 49.7 bits (117), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 46/97 (47%), Gaps = 2/97 (2%)
Query 181 WTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEA 240
W +A L + A V + +LP ++ ++ EW +E+ +L LG + EG +
Sbjct 373 WNIRKCSAAALDILANVFCEELLPHILPLLKELLFHLEWVIKESGILVLGAIAEG-CMQG 431
Query 241 MAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQ 277
M PY+ P L++ +A D VR WTL + A
Sbjct 432 MIPYLPELIPHLIQCLA-DKKALVRSITCWTLSRYAH 467
> pfa:PFF1345w transportin
Length=1147
Score = 49.3 bits (116), Expect = 2e-05, Method: Composition-based stats.
Identities = 25/96 (26%), Positives = 50/96 (52%), Gaps = 2/96 (2%)
Query 179 DSWTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSA 238
+ WT AA+CL + V D IL V+ ++ S +W RE++VL LG + +G
Sbjct 524 NDWTVRKGAALCLDYLSNVYNDEILEFVLPHIEEKLMSDKWNIRESAVLTLGAIAKG-CM 582
Query 239 EAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGK 274
+++P++ + L++ + + +A R + W + +
Sbjct 583 YSLSPFIPKVLEYLIKLLNDEKPLA-RSISCWCVTR 617
> tpv:TP04_0218 transportin
Length=971
Score = 49.3 bits (116), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 32/137 (23%), Positives = 69/137 (50%), Gaps = 16/137 (11%)
Query 169 LNAAEDEDDD---DSWTPAMAAAICLGLCAQVVKDA---ILPLVINFVQLNFGSTEWRRR 222
+ + EDE+ + ++WT +A+ L +Q+ + ++ ++++++Q ST+W +
Sbjct 358 IKSREDEETNTWGNTWTVRKGSALLLDTISQLYGQSNPEVIKILLSYIQEKLDSTDWELK 417
Query 223 EASVLALGCVMEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPI 282
E+ VL LG + +G S + PY+ + L+ VA+D +R + W L + +
Sbjct 418 ESGVLTLGAISKG-SLYTLYPYLPKVIDYLI-VVATDPKPLLRIISCWCLSRFVEW---- 471
Query 283 VLRHLFPPADVSSTTSP 299
LF P ++++ S
Sbjct 472 ----LFLPNNINTYLSK 484
> mmu:240614 Ranbp6, C630001B19, FLJ11120; RAN binding protein
6
Length=1105
Score = 48.1 bits (113), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 69/267 (25%), Positives = 118/267 (44%), Gaps = 40/267 (14%)
Query 34 PQLKVAALRALYHALIFAKENFAKTEERDYIISSVLEAGNAANPSAVQV--AALECIVQI 91
P ++ + RA ++ + N A ++ ++ +L+A N S Q + LE +V+I
Sbjct 195 PAIRTLSARAAATFVLANENNIALFKDFADLLPGILQA---VNDSCYQDDDSVLESLVEI 251
Query 92 SQEYYSFLGAYMPAVGSVTWNALKSSDSAVVIA--ALEVWNALAEEELAIIEGESTRWTL 149
+ +LG Y+ ++ S + + ALEV L+E +++ +
Sbjct 252 ADTVPKYLGPYLEDTLQLSLKLCGDSRLSNLQRQLALEVIVTLSETATPMLKKHT----- 306
Query 150 LNIMKEATPFLLPLLLE-----QLLNAAEDEDDD-DSWTPAMAAAI---CLGLCAQVVKD 200
NI+ +A P +L ++++ +NA E E+DD DS A +A+ GL +VV
Sbjct 307 -NIIAQAVPHILAMMVDLQDDDDWVNADEMEEDDFDSNAVAAESALDRLACGLGGKVV-- 363
Query 201 AILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEAMAPYVARSFPILVEAVAS-- 258
LP+ + S +W+ R A ++AL + EG + PIL E V S
Sbjct 364 --LPMTKEHIMQMLQSPDWKCRHAGLMALSAIGEGCHQQME--------PILDETVNSVL 413
Query 259 ----DASVAVRDSAAWTLGKTAQQHAP 281
D VR +A TLG+ A AP
Sbjct 414 LFLQDPHPRVRAAACTTLGQMATDFAP 440
> bbo:BBOV_II003640 18.m06305; transportin
Length=916
Score = 47.4 bits (111), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 47/203 (23%), Positives = 85/203 (41%), Gaps = 48/203 (23%)
Query 116 SSDSAVVIAALEVWNALAEEELAIIEGESTRWTLLNIMKEATPFLLPLLLEQL------- 168
++D+ V + AL+ W L + L ES L++ ++ P L+P+L+E
Sbjct 279 TADNTVQLEALQFWAQLLKSRLE----ESVNSRLISQLRTHLPQLIPVLIEHTRYSSWDY 334
Query 169 -----------------------------LNAAEDEDD---DDSWTPAMAAAICLGLCAQ 196
+ A EDE+ ++WTP AA+ L +Q
Sbjct 335 MSMDESHFEEDNAAVPDRVEDVPPRPEGEMTADEDEESATWGNNWTPRKGAALALDYISQ 394
Query 197 VV--KDAILPLVINFVQLNFGS-TEWRRREASVLALGCVMEGPSAEAMAPYVARSFPILV 253
V + I+ ++ ++ + ++W +E++VL LG + G AMAPY+ + L+
Sbjct 395 VYGQDNEIVQFLLEHIEKRLANDSDWEMKESAVLVLGAIASG-CMLAMAPYLPKVVEYLI 453
Query 254 EAVASDASVAVRDSAAWTLGKTA 276
E + +R A W L + A
Sbjct 454 E-LTRHPKPLMRSIACWCLARYA 475
> hsa:26953 RANBP6, FLJ54311; RAN binding protein 6
Length=1105
Score = 47.4 bits (111), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 60/223 (26%), Positives = 100/223 (44%), Gaps = 29/223 (13%)
Query 74 AANPSAVQV--AALECIVQISQEYYSFLGAYMPAVGSVTWNALKSSDSAVVIA--ALEVW 129
A N S Q + LE +V+I+ +LG Y+ ++ S + + ALEV
Sbjct 232 AVNDSCYQDDDSVLESLVEIADTVPKYLGPYLEDTLQLSLKLCGDSRLSNLQRQLALEVI 291
Query 130 NALAEEELAIIEGESTRWTLLNIMKEATPFLLPLLL-----EQLLNAAEDEDDD-DSWTP 183
L+E +++ + NI+ +A P +L +++ E +NA E E+DD DS
Sbjct 292 VTLSETATPMLKKHT------NIIAQAVPHILAMMVDLQDDEDWVNADEMEEDDFDSNAV 345
Query 184 AMAAAI---CLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEG--PSA 238
A +A+ GL +VV LP+ + S +W+ R A ++AL + EG
Sbjct 346 AAESALDRLACGLGGKVV----LPMTKEHIMQMLQSPDWKYRHAGLMALSAIGEGCHQQM 401
Query 239 EAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAP 281
E++ S + ++ D VR +A TLG+ A AP
Sbjct 402 ESILDETVNSVLLFLQ----DPHPRVRAAACTTLGQMATDFAP 440
> dre:445499 tnpo2, ik:tdsubc_2a7, tdsubc_2a7, wu:fb01c02, wu:fe01f03,
wu:fe16e12, xx:tdsubc_2a7, zgc:101009; transportin
2 (importin 3, karyopherin beta 2b)
Length=889
Score = 46.2 bits (108), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 46/97 (47%), Gaps = 2/97 (2%)
Query 181 WTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEA 240
W +A L + A V +D +LP ++ ++ +W +E+ +L LG + EG +
Sbjct 372 WNLRKCSAAALDVLANVFRDELLPHLLPVLKELLFHPDWVVKESGILVLGAIAEG-CMQD 430
Query 241 MAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQ 277
M Y+ P LV+ + D VR A WTL + A
Sbjct 431 MVLYLPELIPHLVQCLC-DKKALVRSIACWTLSRYAH 466
> ath:AT2G16960 importin beta-2 subunit family protein
Length=505
Score = 45.4 bits (106), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 36/118 (30%), Positives = 52/118 (44%), Gaps = 13/118 (11%)
Query 166 EQLLNAAEDED--------DDDSWTPAMAAAICLGLCAQVVKDAILPLVINFVQLN---F 214
E LLN E E +D W +A +G+ A V D IL ++ ++ F
Sbjct 8 ETLLNEEEVESQPDIDQAQNDKEWNLRACSAKFIGILANVFGDEILLTLMPLIEAKLSKF 67
Query 215 GSTEWRRREASVLALGCVMEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTL 272
W+ REA+V A G + EG ++ P++A IL + D S VR WTL
Sbjct 68 DDETWKEREAAVFAFGAIAEGCNS-FFYPHLAEIVAIL-RRLLDDQSPLVRRITCWTL 123
> hsa:30000 TNPO2, FLJ12155, IPO3, KPNB2B, TRN2; transportin 2
Length=887
Score = 44.3 bits (103), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 22/60 (36%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query 218 EWRRREASVLALGCVMEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQ 277
EW +E+ +L LG + EG + M PY+ P L++ + SD VR A WTL + A
Sbjct 407 EWVVKESGILVLGAIAEG-CMQGMVPYLPELIPHLIQCL-SDKKALVRSIACWTLSRYAH 464
> xla:495010 hypothetical LOC495010
Length=889
Score = 44.3 bits (103), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 22/60 (36%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query 218 EWRRREASVLALGCVMEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQ 277
EW +E+ +L LG + EG + M PY+ P L++ + SD VR A WTL + A
Sbjct 409 EWVIKESGILVLGAIAEG-CMQGMVPYLPELIPHLIQCL-SDKKALVRSIACWTLSRYAH 466
> mmu:212999 Tnpo2, 1110034O24Rik, AA414969, AI464345, AI852433,
IPO3, Knpb2b, Kpnb2b, MGC6380, TRN2; transportin 2 (importin
3, karyopherin beta 2b)
Length=897
Score = 44.3 bits (103), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 22/60 (36%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query 218 EWRRREASVLALGCVMEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQ 277
EW +E+ +L LG + EG + M PY+ P L++ + SD VR A WTL + A
Sbjct 407 EWVVKESGILVLGAIAEG-CMQGMVPYLPELIPHLIQCL-SDKKALVRSIACWTLSRYAH 464
> xla:495499 tnpo2; transportin 2
Length=890
Score = 44.3 bits (103), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 22/60 (36%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query 218 EWRRREASVLALGCVMEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQ 277
EW +E+ +L LG + EG + M PY+ P L++ + SD VR A WTL + A
Sbjct 410 EWVIKESGILVLGAIAEG-CMQGMVPYLPELIPHLIQCL-SDKKALVRSIACWTLSRYAH 467
> sce:YBR017C KAP104; Kap104p
Length=918
Score = 43.5 bits (101), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 30/113 (26%), Positives = 51/113 (45%), Gaps = 6/113 (5%)
Query 181 WTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEA 240
W +A L + ++ ++ + F++ + GS W REA++LALG + EG +
Sbjct 397 WNLRKCSAATLDVMTNILPHQVMDIAFPFLREHLGSDRWFIREATILALGAMAEG-GMKY 455
Query 241 MAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTA----QQHAPIVLRHLFP 289
+ P LVE + +D VR WTL + + Q H ++ L P
Sbjct 456 FNDGLPALIPFLVEQL-NDKWAPVRKMTCWTLSRFSPWILQDHTEFLIPVLEP 507
> ath:AT4G27640 importin beta-2 subunit family protein
Length=1048
Score = 42.7 bits (99), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 64/258 (24%), Positives = 119/258 (46%), Gaps = 22/258 (8%)
Query 35 QLKVAALRALYHALIFAKENFAKTEERDYIISSVLEAGN---AANPSAVQVAALECIVQI 91
+++VAAL+A+ L F + + RD+I S+L+ A+ V + A E ++
Sbjct 175 RVRVAALKAVGSFLEFTNDGDEVVKFRDFI-PSILDVSRKCIASGEEDVAILAFEIFDEL 233
Query 92 SQEYYSFLGAYMPAV--GSVTWNALKSSDSAVVIAALEVWNALAEEELAIIEGESTRWTL 149
+ LG + A+ S+ + ++ +S+ A+++ + LA+ + ++ +
Sbjct 234 IESPAPLLGDSVKAIVQFSLEVSCNQNLESSTRHQAIQIVSWLAKYKYNSLKKHKLVIPI 293
Query 150 LNIMKEATPFLLPLLLEQLLNAAEDEDDDDSWTPAMAAA-ICLGLCAQVVKDAILPLVIN 208
L +M PLL E +D P A+A + L + K LP V+
Sbjct 294 LQVM-------CPLLAESSDQEDDD-----DLAPDRASAEVIDTLAMNLPKHVFLP-VLE 340
Query 209 FVQLNFGSTEWRRREASVLALGCVMEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSA 268
F ++ ST + REASV ALG + EG + M + I++ A+ D + VR +A
Sbjct 341 FASVHCQSTNLKFREASVTALGVISEG-CFDLMKEKLDTVLNIVLGAL-RDPELVVRGAA 398
Query 269 AWTLGKTAQQHAPIVLRH 286
++ +G+ A+ P +L H
Sbjct 399 SFAIGQFAEHLQPEILSH 416
> dre:100005536 transportin-like
Length=889
Score = 42.7 bits (99), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 47/97 (48%), Gaps = 2/97 (2%)
Query 181 WTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEA 240
W +A L + A V +D +LP ++ ++ +W +E+ +L LG + EG +
Sbjct 372 WNLRKCSAAALDVLANVFRDELLPHLLPLLKGLLFHPDWVIKESGILVLGAIAEG-CMQG 430
Query 241 MAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQ 277
M PY+ P L++ + D VR A WTL + A
Sbjct 431 MVPYLPELIPHLIQCLC-DKKALVRSIACWTLSRYAH 466
> cpv:cgd8_3440 importin beta like ARM repeat alpha superhelix
Length=936
Score = 42.4 bits (98), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 28/124 (22%), Positives = 59/124 (47%), Gaps = 14/124 (11%)
Query 172 AEDEDDD-------DSWTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREA 224
+E DD+ + WT A+A+ L + + D IL ++ ++ W ++E+
Sbjct 382 SESNDDEVELGAWGNQWTVRKASALALDHISVIYGDEILGELLPKIEATLQDPNWEKQES 441
Query 225 SVLALGCVMEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTA-----QQH 279
++L LG + G + ++P++ R LV+ + S++ +R + W + + QQ
Sbjct 442 AILVLGAIARG-CIKGLSPFLPRVLSYLVK-LTSNSKPLIRSISCWCISRFTPWLALQQG 499
Query 280 APIV 283
PI+
Sbjct 500 QPIL 503
> dre:562607 tnpo1; transportin 1
Length=891
Score = 42.0 bits (97), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 29/97 (29%), Positives = 45/97 (46%), Gaps = 2/97 (2%)
Query 181 WTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEA 240
W +A L + A V +D +L ++ ++ EW +E+ +L LG + EG +
Sbjct 374 WNLRKCSAAALDVLANVFRDDLLLHILPLLKELLFHPEWLIKESGILVLGAIAEG-CMQG 432
Query 241 MAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQ 277
M PY+ P LV + SD VR WTL + A
Sbjct 433 MIPYLPELIPHLVLCL-SDKKALVRSITCWTLSRYAH 468
> dre:569455 kpnb3, cb273, wu:fc38a10; karyopherin (importin)
beta 3
Length=1077
Score = 40.8 bits (94), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 72/325 (22%), Positives = 138/325 (42%), Gaps = 46/325 (14%)
Query 15 QQRCNVVLTAVAHGLKD-PDPQLKVAALRALYHALIFAKENFAKTEERDYIISSVLEAGN 73
Q V+ + ++D +PQ++ + RA ++ + N A + ++ +L+A N
Sbjct 163 QHYMEVIKRMLVQCMQDQENPQIRTLSARAAASFILSNEGNTALLKHFSDLLPGILQAVN 222
Query 74 AANPSAVQVAALECIVQISQEYYSFLGAYMPAVGSVTWNALKSSDSAVVIA--ALEVWNA 131
+ + L+ +V+I+ +L + A ++ ++ + ALEV
Sbjct 223 ESCYRGDD-SVLKSLVEIADTAPKYLRPNLEATLQLSLKLCADTNLTNMQRQLALEVIVT 281
Query 132 LAEEELAIIEGESTRWTLLNIMKEATPFLLPLLL-----EQLLNAAEDEDDD-DSWTPAM 185
L+E A++ + NI+ ++ P +L +++ E+ A E EDDD DS A
Sbjct 282 LSETAAAMLRKHT------NIVAQSVPQMLTMMVDLEEDEEWAMADELEDDDFDSNAVAG 335
Query 186 AAA---ICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEG--PSAEA 240
+A I GL ++ ILP++ + + +W+ R A ++AL + EG EA
Sbjct 336 ESALDRIACGLGGKI----ILPMIKQHIMQMLQNPDWKYRHAGLMALSAIGEGCHQQMEA 391
Query 241 MAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPADVSSTTSPN 300
+ + SF +L D VR +A +G+ A AP +
Sbjct 392 ILSEIV-SFVLL---FCQDPHPRVRYAACNAIGQMATDFAPTFQKKFH------------ 435
Query 301 NNGGALLLAIVERLTDQ--PRVAVH 323
++ A+++ + DQ PRV H
Sbjct 436 ---DKVISALLQTMEDQSNPRVQAH 457
> hsa:63897 HEATR6, ABC1, DKFZp686D22141, FLJ22087, MGC148096;
HEAT repeat containing 6
Length=1181
Score = 40.4 bits (93), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 48/210 (22%), Positives = 96/210 (45%), Gaps = 13/210 (6%)
Query 71 AGNAANPSAVQVAALECIVQISQEYYSFLGAYMPAVGSVTWNALKSSDSAVVIAALEVWN 130
AG+ PS +++ AL+ + +++ Y+S AY+ +G V + +D ++ + ++
Sbjct 678 AGSTYEPSPMRLEALQVLTLLARGYFSMTQAYLMELGEVICKCMGEADPSIQLHGAKLLE 737
Query 131 ALAEEELAIIEGESTRWTLLNIMKEATPFLLPLLLEQLLNAAEDEDDDDSWTPAMAAAIC 190
L + + +ST + A FL+ + +LN +S P + A+ C
Sbjct 738 ELGTGLIQQYKPDSTAAP----DQRAPVFLVVMFWTMMLNGPLPRALQNSEHPTLQASAC 793
Query 191 LGLCAQVVKDAI--LP----LVINFVQLNFGSTEWRR-REASVLALGCVMEGPSAEAMAP 243
L + ++ +A LP ++ V L ++ R + A+ ALG + P
Sbjct 794 DAL-SSILPEAFSNLPNDRQMLCITVLLGLNDSKNRLVKAATSRALGVYVLFPCLRQDVI 852
Query 244 YVARSFPILVEAVASDASVAVRDSAAWTLG 273
+VA + ++ ++ D S+ VR AAW+LG
Sbjct 853 FVADAANAILMSL-EDKSLNVRAKAAWSLG 881
> cel:C53D5.6 imb-3; IMportin Beta family member (imb-3)
Length=1092
Score = 40.0 bits (92), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 72/316 (22%), Positives = 128/316 (40%), Gaps = 60/316 (18%)
Query 32 PDPQLKVAALRALYHALIFAKENFAKTEERDYI------ISSVLEAGNAANPSAVQVAAL 85
PD Q+K A+RA+ + FA +N EE+D + + +VL+ N + L
Sbjct 179 PDLQIKATAVRAV---IAFAVDN---DEEKDVVRLMTSLVPNVLQVCNETSDEDDSDGPL 232
Query 86 ECIVQISQEYYSFLGAYMPAVGSVTWN--ALKSSDSAVVIAALEVWNALAEEELAIIEGE 143
+++ L +M V VT K + V A+EV + E + +G
Sbjct 233 GEFAELASSLPKCLNTHMSQVLQVTLAIAGNKEKNEMVRQNAIEVICSYME---SAPKG- 288
Query 144 STRWTLLNIMKEATPFLLPLLLEQLLNAAEDEDDD--DSW------------TPAMAAAI 189
+K+ P L +LE LL+ + DDD + W P +A +
Sbjct 289 ---------LKKYAPGALGPILETLLSCMTEMDDDVLNEWLNEIEEEEDYEDIPIIAESA 339
Query 190 CLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEAMAPYVARSF 249
+ + +LP+ + V+ S +W+ + A++ A V EG +M P++ +
Sbjct 340 IDRVACCINGKVMLPVFLPLVEKLLTSEDWKMKHAALRAFSAVGEG-CQRSMEPHIEQIM 398
Query 250 PILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPADVSSTTSPNNNGGALLLA 309
+ V +DA V+ +A +G+ + AP + + A++ A
Sbjct 399 AHITNYV-NDAHPRVQYAACNAIGQMSSDFAPTLQKKCH---------------AAVIPA 442
Query 310 IVERL--TDQPRVAVH 323
++E L TD PRV H
Sbjct 443 LLESLDRTDVPRVCAH 458
> hsa:3843 IPO5, DKFZp686O1576, FLJ43041, IMB3, KPNB3, MGC2068,
RANBP5; importin 5
Length=1115
Score = 38.5 bits (88), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 63/284 (22%), Positives = 117/284 (41%), Gaps = 35/284 (12%)
Query 15 QQRCNVVLTAVAHGLKDPD-PQLKVAALRALYHALIFAKENFAKTEERDYIISSVLEAGN 73
Q +V+ + ++D + P ++ + RA ++ + N A + ++ L+A
Sbjct 185 QHYLDVIKRMLVQCMQDQEHPSIRTLSARATAAFILANEHNVALFKHFADLLPGFLQA-- 242
Query 74 AANPSAVQV--AALECIVQISQEYYSFLGAYMPAV--------GSVTWNALKSSDSAVVI 123
N S Q + L+ +V+I+ +L ++ A G + N ++
Sbjct 243 -VNDSCYQNDDSVLKSLVEIADTVPKYLRPHLEATLQLSLKLCGDTSLNNMQRQ------ 295
Query 124 AALEVWNALAEEELAIIEGESTRWTLLNIMKEATPFLLPLLL-----EQLLNAAEDEDDD 178
ALEV L+E A++ + NI+ + P +L +++ E NA E EDDD
Sbjct 296 LALEVIVTLSETAAAMLRKHT------NIVAQTIPQMLAMMVDLEEDEDWANADELEDDD 349
Query 179 -DSWTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPS 237
DS A +A+ C K +LP++ + + +W+ R A ++AL + EG
Sbjct 350 FDSNAVAGESALDRMACGLGGK-LVLPMIKEHIMQMLQNPDWKYRHAGLMALSAIGEGCH 408
Query 238 AEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAP 281
+ + V D VR +A +G+ A AP
Sbjct 409 QQMEG--ILNEIVNFVLLFLQDPHPRVRYAACNAVGQMATDFAP 450
> tgo:TGME49_016590 transportin, putative
Length=945
Score = 38.5 bits (88), Expect = 0.037, Method: Compositional matrix adjust.
Identities = 28/98 (28%), Positives = 51/98 (52%), Gaps = 2/98 (2%)
Query 179 DSWTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSA 238
D W+ +A+ L A V ++A+LP V+ ++ + W RREA+VLALG + +G
Sbjct 467 DGWSVRKGSALALDHIASVYREAVLPEVLPLIEASLVDANWERREAAVLALGALAQG-CQ 525
Query 239 EAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTA 276
+++ PY+ L+ + D +R + W + + A
Sbjct 526 DSLEPYLPNVLQFLLN-LCDDPKPLLRSISCWCVSRYA 562
> cel:R06A4.4 imb-2; IMportin Beta family member (imb-2)
Length=883
Score = 38.1 bits (87), Expect = 0.060, Method: Compositional matrix adjust.
Identities = 23/100 (23%), Positives = 44/100 (44%), Gaps = 2/100 (2%)
Query 181 WTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEA 240
W +A L + A + +L + ++ + W +E+ +LALG + EG +
Sbjct 363 WNIRRCSAASLDVLASIFGKDLLDKLFPLLKDTLMNDNWLVKESGILALGAIAEG-CMDG 421
Query 241 MAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHA 280
+ P++ P ++ A+ D VR WTL + + A
Sbjct 422 VVPHLGELIPFML-AMMFDKKPLVRSITCWTLSRYSSHIA 460
> xla:444842 ipo5-a, MGC132286, ipo5, kap_beta_3, ranbp5; importin
5
Length=1094
Score = 36.6 bits (83), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 50/210 (23%), Positives = 92/210 (43%), Gaps = 23/210 (10%)
Query 83 AALECIVQISQEYYSFLGAYMPAVGSVTWN--ALKSSDSAVVIAALEVWNALAEEELAII 140
+ L+ +V+I+ FL ++ A ++ A +S + A+EV L+E A++
Sbjct 232 SVLKSLVEIADTVPKFLRPHLEATLQLSLKLFADRSLSNMQRQLAMEVIVTLSETAAAML 291
Query 141 EGESTRWTLLNIMKEATPFLLPLLLE-----QLLNAAEDEDDD-DSWTPAMAAAI---CL 191
++ I+ +A P +L ++++ NA E EDDD DS A +A+
Sbjct 292 RKHTS------IVAQAIPQMLAMMVDLEDDDDWSNADELEDDDFDSNAVAGESALDRMAC 345
Query 192 GLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEAMAPYVARSFPI 251
GL ++V LP++ + + +W+ R A ++AL + EG + +
Sbjct 346 GLGGKIV----LPMIKEHIMQMLQNPDWKYRHAGLMALSAIGEGCHQQMEG--ILNEMVN 399
Query 252 LVEAVASDASVAVRDSAAWTLGKTAQQHAP 281
V D VR +A +G+ A AP
Sbjct 400 FVLLFLQDPHPRVRYAACNAIGQMATDFAP 429
> mmu:70572 Ipo5, 1110011C18Rik, 5730478E03Rik, AA409333, C76941,
IMB3, Kpnb3, Ranbp5; importin 5
Length=1097
Score = 35.4 bits (80), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 61/284 (21%), Positives = 116/284 (40%), Gaps = 35/284 (12%)
Query 15 QQRCNVVLTAVAHGLKDPD-PQLKVAALRALYHALIFAKENFAKTEERDYIISSVLEAGN 73
Q +V+ + ++D + P ++ + RA ++ + N A + ++ L+A
Sbjct 167 QHYLDVIKRMLVQCMQDQEHPSIRTLSARATAAFILANEHNVALFKHFADLLPGFLQA-- 224
Query 74 AANPSAVQV--AALECIVQISQEYYSFLGAYMPAV--------GSVTWNALKSSDSAVVI 123
N S Q + L+ +V+I+ +L ++ A G N ++
Sbjct 225 -VNDSCYQNDDSVLKSLVEIADTVPKYLRPHLEATLQLSLKLCGDTNLNNMQRQ------ 277
Query 124 AALEVWNALAEEELAIIEGESTRWTLLNIMKEATPFLLPLLL-----EQLLNAAEDEDDD 178
ALEV L+E A++ ++ ++ + P +L +++ E NA E EDDD
Sbjct 278 LALEVIVTLSETAAAMLRKHTS------LIAQTIPQMLAMMVDLEEDEDWANADELEDDD 331
Query 179 -DSWTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPS 237
DS A +A+ C K +LP++ + + +W+ R A ++AL + EG
Sbjct 332 FDSNAVAGESALDRMACGLGGK-LVLPMIKEHIMQMLQNPDWKYRHAGLMALSAIGEGCH 390
Query 238 AEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAP 281
+ + V D VR +A +G+ A AP
Sbjct 391 QQMEG--ILNEIVNFVLLFLQDPHPRVRYAACNAVGQMATDFAP 432
> xla:444846 ipo5-b, ranbp5, ranbp6; importin 5
Length=1094
Score = 35.0 bits (79), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 42/166 (25%), Positives = 73/166 (43%), Gaps = 21/166 (12%)
Query 125 ALEVWNALAEEELAIIEGESTRWTLLNIMKEATPFLLPLLLE-----QLLNAAEDEDDD- 178
A+EV L+E A++ ++ I+ +A P +L ++++ NA E EDDD
Sbjct 276 AMEVIVTLSETAAAMLRKHTS------IVAQAIPQMLAMMVDLEDDDDWSNADELEDDDF 329
Query 179 DSWTPAMAAAI---CLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEG 235
DS A +A+ GL ++V LP++ + + +W+ R A ++AL + EG
Sbjct 330 DSNAVAGESALDRMACGLGGKIV----LPMIKEHIMQMLQNPDWKYRHAGLMALSAIGEG 385
Query 236 PSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAP 281
+ + V D VR +A +G+ A AP
Sbjct 386 CHQQMEG--ILNEMVNFVLLFLQDPHPRVRYAACNAIGQMATDFAP 429
> cel:Y48G1A.5 xpo-2; eXPOrtin (nuclear export receptor) family
member (xpo-2)
Length=938
Score = 33.9 bits (76), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 40/149 (26%), Positives = 61/149 (40%), Gaps = 18/149 (12%)
Query 86 ECIVQISQEYYSFLGAYMPAVGSVTWNALKSSD-----SAVVIAALEVWNALAEEELAII 140
E SQ Y + ++P + WN LKS+ +V AALE + +++ +
Sbjct 267 EIFTLYSQRYEEEISEFVPDIILAVWNLLKSTGPDTRYDTMVCAALEFLSMVSQRQY--Y 324
Query 141 EGESTRWTLLNIMKEATPFLLPLLLEQLLNAAEDEDDD---------DSWTPAMAAA-IC 190
EG T +L + E LL +Q + EDE D D T A +
Sbjct 325 EGHFTGEGVLKTLAENVCVQNLLLRQQDMELFEDEPLDYMKRDIEGTDVGTRRRGAIDLA 384
Query 191 LGLCAQVVKDAILPLVINFVQLNFGSTEW 219
GLC + + +LP + VQ GS +W
Sbjct 385 RGLCRR-FEAQMLPCLGEIVQNLLGSGDW 412
> ath:AT1G72440 EDA25; EDA25 (embryo sac development arrest 25);
K14832 ribosome biogenesis protein MAK21
Length=1043
Score = 33.5 bits (75), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 33/65 (50%), Gaps = 5/65 (7%)
Query 15 QQRCNVVLTAVAHGLKDPDPQLKVAALRALYHALIFAKENFAKTEERDYIISSVLEAGNA 74
+QR +TA+ KD P+LK AL+ +Y L E +ER ++S V + G+
Sbjct 302 KQRYERFVTALDESSKDMLPELKDKALKTIYFMLTSKSE-----QERKLLVSLVNKLGDP 356
Query 75 ANPSA 79
N SA
Sbjct 357 QNKSA 361
> dre:100334527 si:dkey-245p14.6
Length=972
Score = 33.5 bits (75), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 29/99 (29%), Positives = 43/99 (43%), Gaps = 15/99 (15%)
Query 213 NFGSTEWRRREASVLALGCVMEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTL 272
N S WR RE+S LAL ++ G A+ + ++ + L V D +VR +A TL
Sbjct 295 NLTSNMWRVRESSCLALNDLIRGRQADEIIDRLSEIWETLFR-VLDDIKESVRKAADLTL 353
Query 273 --------------GKTAQQHAPIVLRHLFPPADVSSTT 297
G TAQ+ ++L L VS+ T
Sbjct 354 KTLSKVCVRMCESTGATAQRTVAVLLPTLLDKGIVSNVT 392
> sce:YIL128W MET18, MMS19; DNA repair and TFIIH regulator, required
for both nucleotide excision repair (NER) and RNA polymerase
II (RNAP II) transcription; involved in telomere maintenance;
K15075 DNA repair/transcription protein MET18/MMS19
Length=1032
Score = 33.5 bits (75), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 17/60 (28%), Positives = 33/60 (55%), Gaps = 1/60 (1%)
Query 228 ALGCVMEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHL 287
AL V++ ++++ P++ FP+L++A+ VR SA TL T +H ++ H+
Sbjct 891 ALSLVLKHTPSQSVGPFINDLFPLLLQALDM-PDPEVRVSALETLKDTTDKHHTLITEHV 949
> ath:AT1G47380 protein phosphatase 2C-related / PP2C-related
Length=428
Score = 33.5 bits (75), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 29/103 (28%), Positives = 48/103 (46%), Gaps = 5/103 (4%)
Query 59 EERDYIISSVLEAGNAANPSAVQVAALEC---IVQISQEYYSF-LGAYMPAVGSVTWNAL 114
EERD + +S E G ++ L C + +S+ +G Y+ V V L
Sbjct 174 EERDRVTASGGEVGRLNTGGGTEIGPLRCWPGGLCLSRSIGDLDVGEYIVPVPYVKQVKL 233
Query 115 KSSDSAVVIAALEVWNAL-AEEELAIIEGESTRWTLLNIMKEA 156
S+ ++I++ VW+A+ AEE L G + +I+KEA
Sbjct 234 SSAGGRLIISSDGVWDAISAEEALDCCRGLPPESSAEHIVKEA 276
> ath:AT1G50030 TOR; TOR (TARGET OF RAPAMYCIN); 1-phosphatidylinositol-3-kinase/
protein binding; K07203 FKBP12-rapamycin
complex-associated protein
Length=2454
Score = 33.5 bits (75), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 25/44 (56%), Gaps = 0/44 (0%)
Query 240 AMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIV 283
AM Y+ P++VEA+ A+VA R+ A TLG+ Q +V
Sbjct 734 AMRQYIPELMPLIVEALMDGAAVAKREVAVSTLGQVVQSTGYVV 777
> dre:563777 similar to Proteasome-associated protein ECM29 homolog;
K11886 proteasome component ECM29
Length=1867
Score = 33.5 bits (75), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 29/99 (29%), Positives = 43/99 (43%), Gaps = 15/99 (15%)
Query 213 NFGSTEWRRREASVLALGCVMEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTL 272
N S WR RE+S LAL ++ G A+ + ++ + L V D +VR +A TL
Sbjct 1154 NLTSNMWRVRESSCLALNDLIRGRQADEIIDRLSEIWETLFR-VLDDIKESVRKAADLTL 1212
Query 273 --------------GKTAQQHAPIVLRHLFPPADVSSTT 297
G TAQ+ ++L L VS+ T
Sbjct 1213 KTLSKVCVRMCESTGATAQRTVAVLLPTLLDKGIVSNVT 1251
Lambda K H
0.317 0.131 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 15956348688
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40