bitscore colors: <40, 40-50 , 50-80, 80-200, >200
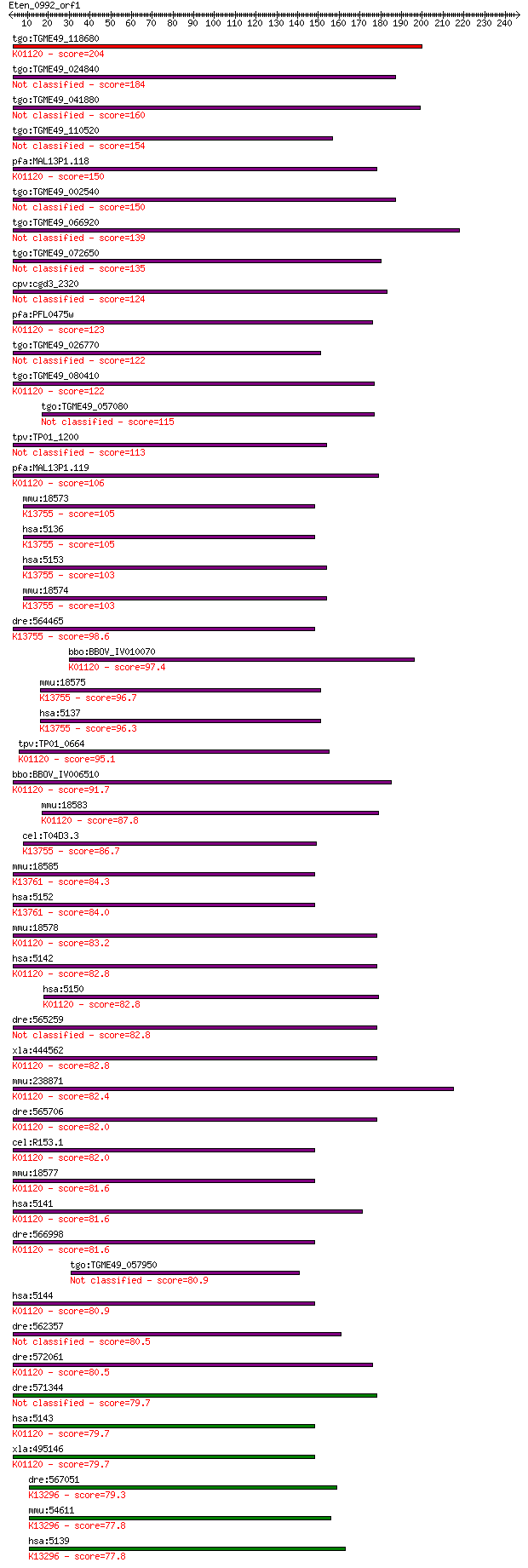
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0992_orf1
Length=246
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_118680 3',5'--cyclic-nucleotide phosphodiesterase, ... 204 3e-52
tgo:TGME49_024840 cAMP phosphodiesterase, putative (EC:3.1.4.1... 184 2e-46
tgo:TGME49_041880 3', 5'-cyclic nucleotide phosphodiesterase, ... 160 3e-39
tgo:TGME49_110520 calcium/calmodulin-dependent 3', 5'-cyclic n... 154 2e-37
pfa:MAL13P1.118 3',5'-cyclic nucleotide phosphodiesterase (EC:... 150 4e-36
tgo:TGME49_002540 3',5'-cyclic nucleotide phosphodiesterase, p... 150 6e-36
tgo:TGME49_066920 phosphodiesterase, putative (EC:3.1.4.17) 139 1e-32
tgo:TGME49_072650 calcium/calmodulin-dependent 3',5'-cyclic nu... 135 1e-31
cpv:cgd3_2320 cGMP phosphodiesterase A4 124 4e-28
pfa:PFL0475w PDE1; cGMP-specific phosphodiesterase (EC:3.1.4.1... 123 7e-28
tgo:TGME49_026770 cGMP-inhibited 3',5'-cyclic phosphodiesteras... 122 9e-28
tgo:TGME49_080410 3',5'-cyclic phosphodiesterase, putative (EC... 122 2e-27
tgo:TGME49_057080 cAMP-specific 3',5'-cyclic phosphodiesterase... 115 2e-25
tpv:TP01_1200 hypothetical protein 113 6e-25
pfa:MAL13P1.119 calcium/calmodulin-dependent 3',5'-cyclic nucl... 106 8e-23
mmu:18573 Pde1a, AI987702, AW125737, MGC116577; phosphodiester... 105 1e-22
hsa:5136 PDE1A, HCAM1, HSPDE1A, MGC26303; phosphodiesterase 1A... 105 2e-22
hsa:5153 PDE1B, PDE1B1, PDES1B; phosphodiesterase 1B, calmodul... 103 7e-22
mmu:18574 Pde1b, 63kDa, Pde1b1; phosphodiesterase 1B, Ca2+-cal... 103 7e-22
dre:564465 pde1a; phosphodiesterase 1A, calmodulin-dependent; ... 98.6 2e-20
bbo:BBOV_IV010070 23.m06154; 3'5'-cyclic nucleotide phosphodie... 97.4 4e-20
mmu:18575 Pde1c; phosphodiesterase 1C (EC:3.1.4.17); K13755 ca... 96.7 7e-20
hsa:5137 PDE1C, Hcam3; phosphodiesterase 1C, calmodulin-depend... 96.3 8e-20
tpv:TP01_0664 3',5' cyclic nucleotide phosphodiesterase; K0112... 95.1 2e-19
bbo:BBOV_IV006510 23.m05980; 3'5'-cyclic nucleotide phosphodie... 91.7 2e-18
mmu:18583 Pde7a, AU015378, AW047537; phosphodiesterase 7A (EC:... 87.8 3e-17
cel:T04D3.3 pde-1; PhosphoDiEsterase family member (pde-1); K1... 86.7 7e-17
mmu:18585 Pde9a, PDE9A1; phosphodiesterase 9A (EC:3.1.4.35); K... 84.3 3e-16
hsa:5152 PDE9A, FLJ90181, HSPDE9A2; phosphodiesterase 9A (EC:3... 84.0 5e-16
mmu:18578 Pde4b, Dpde4, R74983, dunce; phosphodiesterase 4B, c... 83.2 8e-16
hsa:5142 PDE4B, DKFZp686F2182, DPDE4, MGC126529, PDE4B5, PDEIV... 82.8 9e-16
hsa:5150 PDE7A, HCP1, PDE7; phosphodiesterase 7A (EC:3.1.4.17)... 82.8 1e-15
dre:565259 pde4c, im:7160317, si:dkey-149i17.5; phosphodiester... 82.8 1e-15
xla:444562 pde4b, MGC83972; phosphodiesterase 4B, cAMP-specifi... 82.8 1e-15
mmu:238871 Pde4d, 9630011N22Rik, Dpde3; phosphodiesterase 4D, ... 82.4 2e-15
dre:565706 cAMP-specific 3,5-cyclic phosphodiesterase 4B-like;... 82.0 2e-15
cel:R153.1 pde-4; PhosphoDiEsterase family member (pde-4); K01... 82.0 2e-15
mmu:18577 Pde4a, D9Ertd60e, Dpde2; phosphodiesterase 4A, cAMP ... 81.6 2e-15
hsa:5141 PDE4A, DPDE2, PDE4, PDE46; phosphodiesterase 4A, cAMP... 81.6 2e-15
dre:566998 novel protein similar to vertebrate phosphodiestera... 81.6 2e-15
tgo:TGME49_057950 3'5'-cyclic nucleotide phosphodiesterase dom... 80.9 4e-15
hsa:5144 PDE4D, DKFZp686M11213, DPDE3, FLJ97311, HSPDE4D, PDE4... 80.9 4e-15
dre:562357 phosphodiesterase 9A-like 80.5 5e-15
dre:572061 pde4a; phosphodiesterase 4A, cAMP-specific; K01120 ... 80.5 5e-15
dre:571344 fc68c05, si:ch211-255d18.8, wu:fc68c05; si:ch211-25... 79.7 8e-15
hsa:5143 PDE4C, DPDE1, MGC126222; phosphodiesterase 4C, cAMP-s... 79.7 8e-15
xla:495146 hypothetical LOC495146; K01120 3',5'-cyclic-nucleot... 79.7 1e-14
dre:567051 si:dkey-48h7.2; K13296 cGMP-inhibited 3',5'-cyclic ... 79.3 1e-14
mmu:54611 Pde3a, A930022O17Rik, C87899; phosphodiesterase 3A, ... 77.8 3e-14
hsa:5139 PDE3A, CGI-PDE; phosphodiesterase 3A, cGMP-inhibited ... 77.8 3e-14
> tgo:TGME49_118680 3',5'--cyclic-nucleotide phosphodiesterase,
putative (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=812
Score = 204 bits (518), Expect = 3e-52, Method: Compositional matrix adjust.
Identities = 93/197 (47%), Positives = 130/197 (65%), Gaps = 0/197 (0%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H LTF+VL CN+FA L+ +E VR+NMI LILATDMK+HF ++R R RQ P F
Sbjct 447 HSALTFRVLGRHDCNVFAYLSDEEFKHVRANMIDLILATDMKTHFDFLSRFRNRRQAPGF 506
Query 63 DHRKQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPL 122
+ K +D WL E+C+RA+D+GH++ WDQHFEW+SRV EFYLQG+EE R G +SPL
Sbjct 507 NFHKVAEDQWLAAEICIRASDLGHAMTEWDQHFEWASRVVTEFYLQGEEERRLGLHVSPL 566
Query 123 CDRDLHSSMARNQVGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDANISRWQQLQDIGV 182
CDRD H S ++ Q GFL ++VKPL EL D+++ + + + N+ RW+ + GV
Sbjct 567 CDRDDHPSFSKCQHGFLGYVVKPLIAELGECDTLQRIIPQLMQNLKENMERWEAFTNEGV 626
Query 183 DVTFSPVVMACEDRLKG 199
+V S +A + + +G
Sbjct 627 NVVLSSAALAYDIKGQG 643
> tgo:TGME49_024840 cAMP phosphodiesterase, putative (EC:3.1.4.17
1.1.99.3)
Length=1324
Score = 184 bits (467), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 85/184 (46%), Positives = 117/184 (63%), Gaps = 0/184 (0%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H LTF+ L + NIFA L + +R +I ILATDMK HF +++R R R +P F
Sbjct 817 HSCLTFRTLENKDRNIFACLDESSFRYIRQYVIECILATDMKQHFESLSRFRIRRNSPEF 876
Query 63 DHRKQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPL 122
++ K +D W + MC++ ADIGHS + W+QHFEWSSRV EFYLQGDEE +SPL
Sbjct 877 NYVKNVEDRWFVARMCVKVADIGHSCVPWNQHFEWSSRVVEEFYLQGDEERSRNMPVSPL 936
Query 123 CDRDLHSSMARNQVGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDANISRWQQLQDIGV 182
CDRD H+ MA++Q GFLE +VKPL E+ +D + ++ I+ N +WQ+LQ+ G
Sbjct 937 CDRDKHADMAKSQGGFLEFVVKPLIKEIDEIDPFGRVRSDILSHIEYNEKKWQELQESGT 996
Query 183 DVTF 186
+V
Sbjct 997 EVVL 1000
> tgo:TGME49_041880 3', 5'-cyclic nucleotide phosphodiesterase,
putative (EC:2.1.1.43 3.1.4.17)
Length=1281
Score = 160 bits (406), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 79/201 (39%), Positives = 113/201 (56%), Gaps = 6/201 (2%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H LTFK+L+ CNIF L ++ VR+ MI LILATDMK HF +++ R R +P F
Sbjct 937 HACLTFKILARPECNIFTHLPPEDFQIVRNRMIDLILATDMKMHFEMVSKFRLRRNSPDF 996
Query 63 DHRKQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPL 122
R DD W+++ MC++ D+ H +LSWD H WS R +EFY QGD E GR I+P+
Sbjct 997 -CRTNDDDVWMLLRMCIKGGDLSHGLLSWDSHVSWSYRAASEFYEQGDLELAQGRTITPM 1055
Query 123 CDRDLHSSMARNQVGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDANISRWQQLQDI-- 180
DR H+ + Q GFL ++ PL+ E+ +VD+ + AN RW+QLQ
Sbjct 1056 FDRRKHADFPKGQEGFLRFIIIPLYEEIAAVDTTDVVQTCCLQNAVANSERWRQLQSSPE 1115
Query 181 ---GVDVTFSPVVMACEDRLK 198
+D ++ +DRL+
Sbjct 1116 EFEAIDAKHQAIICVLKDRLE 1136
> tgo:TGME49_110520 calcium/calmodulin-dependent 3', 5'-cyclic
nucleotide phosphodiesterase, putative (EC:3.4.21.69 3.1.4.17)
Length=1531
Score = 154 bits (390), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 69/155 (44%), Positives = 101/155 (65%), Gaps = 1/155 (0%)
Query 3 HDVLTFKVLSHTS-CNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPH 61
H L F++LS N+F+ L ++ R N+I+LILATD+K HF I+R R R +P
Sbjct 1290 HSALLFRILSEIPDSNVFSGLPQETFRIARQNIITLILATDIKQHFETISRFRLRRNSPE 1349
Query 62 FDHRKQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISP 121
F+ K+ +D WL+ +M + ADI H+ ++WD HF WS +V AEFY QGD E R G +SP
Sbjct 1350 FNFLKKEEDDWLVRKMIFKIADISHATVAWDAHFFWSCKVNAEFYAQGDAEVRLGLPVSP 1409
Query 122 LCDRDLHSSMARNQVGFLEHLVKPLFVELHSVDSM 156
LCDR+ H M ++QV FL +V+PL EL +++++
Sbjct 1410 LCDREKHFEMGKSQVAFLNFVVEPLLRELEAIEAL 1444
> pfa:MAL13P1.118 3',5'-cyclic nucleotide phosphodiesterase (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=1139
Score = 150 bits (379), Expect = 4e-36, Method: Composition-based stats.
Identities = 69/175 (39%), Positives = 108/175 (61%), Gaps = 0/175 (0%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H +TFK+L CNI + ++ + +RS +I LIL+TDMK HF I++ R R+N F
Sbjct 920 HASITFKILQLNQCNILKNFSEKDFRMMRSYIIELILSTDMKHHFEIISKFRIRRENEDF 979
Query 63 DHRKQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPL 122
D+ K DD ++ +M +++ADI H +SW +H+ W RV +EFY QGDEE ++ +SPL
Sbjct 980 DYIKNSDDLLILTKMIIKSADISHGSVSWSEHYCWCQRVLSEFYTQGDEELKNKMPLSPL 1039
Query 123 CDRDLHSSMARNQVGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDANISRWQQL 177
CDR H+ + ++Q+ FL+ +V PLF EL +D+ K + +++N W L
Sbjct 1040 CDRTKHNEVCKSQITFLKFVVMPLFEELSHIDNNKFIKSFCLKRLNSNCIMWDTL 1094
> tgo:TGME49_002540 3',5'-cyclic nucleotide phosphodiesterase,
putative (EC:3.1.4.17 1.6.99.5)
Length=1670
Score = 150 bits (378), Expect = 6e-36, Method: Compositional matrix adjust.
Identities = 74/184 (40%), Positives = 106/184 (57%), Gaps = 1/184 (0%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H LTFK L C+IF SL + VRS +I LILATDMK+HF ++R R R F
Sbjct 1411 HSCLTFKTLELPDCDIFFSLRTKDYHMVRSLIIDLILATDMKNHFETVSRFRVRRNALDF 1470
Query 63 DHRKQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPL 122
D + +D W +++ M+ AD+ H + W QHF+W R++ EFY QGDEE +SPL
Sbjct 1471 DLSSE-EDFWFAVKIIMKCADLSHCSVPWSQHFQWCQRLSVEFYDQGDEEVARHLPMSPL 1529
Query 123 CDRDLHSSMARNQVGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDANISRWQQLQDIGV 182
CDR+ HS +A++Q+GF+ + PLF EL ++D + ++ + N S W+ L V
Sbjct 1530 CDREKHSEVAKSQLGFMSFVAVPLFEELMAIDGTGNIEKYCISVMKTNASHWEALSSAAV 1589
Query 183 DVTF 186
V
Sbjct 1590 PVPL 1593
> tgo:TGME49_066920 phosphodiesterase, putative (EC:3.1.4.17)
Length=1065
Score = 139 bits (349), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 81/224 (36%), Positives = 111/224 (49%), Gaps = 9/224 (4%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H L F+ L N+F + ++ VRS +I ILATDMK HF I+R R F
Sbjct 687 HCCLCFRTLQKEENNVFYYVDNEQFRFVRSKLIEAILATDMKQHFETISRFRLRTSADDF 746
Query 63 DHRKQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPL 122
D K DD W +++M ++ ADI H L WD HF S + EFY QGDEE R G +S L
Sbjct 747 DVAKHVDDRWAVLKMFVKMADISHVALPWDMHFRLSCDIVEEFYQQGDEELRRGMPLSQL 806
Query 123 CDRDLHSSMARNQVGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDANISRWQQLQDIGV 182
CDR H+ M ++Q GF+E L KPL L DS E ++ N+ RW+Q+ V
Sbjct 807 CDRSKHNEMPKSQEGFIEFLAKPLVGVLIEGDSTGTIKTEVSEQMERNLLRWKQMVADQV 866
Query 183 DVTFSPV---------VMACEDRLKGKDHVIDVSLITETRCRCP 217
V P+ + RL G + V I+++ + P
Sbjct 867 VVPLRPLGPRTQVRTFTASAVRRLAGVESVQAAPTISQSVIKWP 910
> tgo:TGME49_072650 calcium/calmodulin-dependent 3',5'-cyclic
nucleotide phosphodiesterase, putative (EC:3.1.4.17)
Length=1320
Score = 135 bits (340), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 71/177 (40%), Positives = 95/177 (53%), Gaps = 0/177 (0%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H LTF LSH NIF + +VR I LIL+TDM HF + R+ A R+NP F
Sbjct 1136 HACLTFYTLSHRKSNIFRGKTVTQYRDVRKKAIELILSTDMSLHFEFLTRVAARRENPDF 1195
Query 63 DHRKQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPL 122
+ + +D L++ + ++ AD+GH L W+ H WS+R+ EF QGDEE R G +SPL
Sbjct 1196 NFVDREEDRALVLSLAIKVADLGHCALDWEAHTLWSARLKEEFLAQGDEELRLGLPVSPL 1255
Query 123 CDRDLHSSMARNQVGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDANISRWQQLQD 179
CDR +AR+Q F+E L PL L +V V NIS W+ QD
Sbjct 1256 CDRTQQGQLARSQANFIEFLFLPLAQHLAAVAPDDRFEATVVTRARQNISMWEGAQD 1312
> cpv:cgd3_2320 cGMP phosphodiesterase A4
Length=997
Score = 124 bits (310), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 67/183 (36%), Positives = 103/183 (56%), Gaps = 5/183 (2%)
Query 3 HDVLTFKVLSHT---SCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQN 59
H LTF+++ + + + S + E R +I LIL TDM HF ++R R RQ
Sbjct 798 HASLTFRIIYYYGGGAADFLQSWDAELHKEFRRIVIELILETDMHRHFECVSRFRVRRQA 857
Query 60 PHFDHRKQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGI 119
+D D ++ C++AADIGH L W+QH++W V EF+LQGDEE I
Sbjct 858 VDWDPYGDAQDRLMLARTCLKAADIGHGALKWNQHYKWCRSVVEEFFLQGDEEKALSLPI 917
Query 120 SPLCDRDLHSSMARNQVGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDANISRWQQLQD 179
SP+CDR+ + + ++QVGFL + PLF EL VD ++G + ++ I NI+ W+ + +
Sbjct 918 SPICDRE-STDVPKSQVGFLNFVCLPLFQELCYVD-VEGDVRRCIDRILENINNWEDIAE 975
Query 180 IGV 182
G+
Sbjct 976 AGI 978
> pfa:PFL0475w PDE1; cGMP-specific phosphodiesterase (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=954
Score = 123 bits (308), Expect = 7e-28, Method: Composition-based stats.
Identities = 62/173 (35%), Positives = 96/173 (55%), Gaps = 1/173 (0%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H L FK L + + NIF + + N+I IL+TDMK+HF I+ R +++ +
Sbjct 753 HCSLLFKTLKNPNYNIFEHYPYHIFISCKKNIIKAILSTDMKNHFEYISDFRTSKEFIDY 812
Query 63 DHRKQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPL 122
D+ D W I + ++A+DIGHS L W++H EW+ ++ EFYLQG E S L
Sbjct 813 DNLSN-DQIWQIFCLILKASDIGHSTLEWNKHLEWTLKINEEFYLQGLLEKSLNIQNSFL 871
Query 123 CDRDLHSSMARNQVGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDANISRWQ 175
CD + + +A +Q+ FL+HL PLF EL+ + + + I+ NI RW+
Sbjct 872 CDINTMNKLALSQIDFLKHLCIPLFNELNYICKNNDVYTHCIQPIENNIERWE 924
> tgo:TGME49_026770 cGMP-inhibited 3',5'-cyclic phosphodiesterase,
putative (EC:3.1.4.17)
Length=995
Score = 122 bits (307), Expect = 9e-28, Method: Compositional matrix adjust.
Identities = 62/160 (38%), Positives = 93/160 (58%), Gaps = 12/160 (7%)
Query 3 HDVLTFKVLS-HTSC----------NIFASLAKDEILEVRSNMISLILATDMKSHFGAIN 51
H +TF+ L+ +TS ++ + E R ++I LILATDM HF I+
Sbjct 368 HAYITFRTLTCYTSTTDTNGQGCEGSVLKGITPAEYRYFRKHLIELILATDMSQHFSTIS 427
Query 52 RLRAARQNPHFDHRKQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDE 111
+R R++ F+ +D W++ ++CM+ DIGH+ L WDQH++WS RVT EF LQGD
Sbjct 428 TVRVRRESRTFNFITNEEDRWMMKKLCMKIGDIGHAALDWDQHYKWSMRVTEEFLLQGDA 487
Query 112 EARHGRGISPLCDRDLHS-SMARNQVGFLEHLVKPLFVEL 150
E + G +SPLCDR+ + ++Q F+ +LV PL EL
Sbjct 488 ELKAGLPVSPLCDRNAPEIELHKSQSSFINYLVVPLINEL 527
> tgo:TGME49_080410 3',5'-cyclic phosphodiesterase, putative (EC:3.1.4.17
1.6.5.3); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=1085
Score = 122 bits (305), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 69/181 (38%), Positives = 97/181 (53%), Gaps = 7/181 (3%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H LTFK+L + L ++ R +I LI++TDMK+HF I R RQ F
Sbjct 886 HASLTFKLLLDRRLGLLNHLHREAYRSFRMTVIELIMSTDMKTHFEQIANFRVRRQKEEF 945
Query 63 DHRKQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPL 122
D DD +M M +++ADIGH L W H W + V EFY QGDEE R G +S L
Sbjct 946 DPINNYDDRQKVMSMIIKSADIGHGTLPWADHERWCNLVVQEFYEQGDEEKRLGLPVSFL 1005
Query 123 CDRDLHS-SMARNQVGFLEHLVKPLFVELHSVDSMKGAFQE------AVNTIDANISRWQ 175
CDRD H ++QVGFL+ +VKPL+ EL ++++ + + + NIS W+
Sbjct 1006 CDRDQHDREFFKSQVGFLDFVVKPLYEELKALETQLNLNPQLPIENICLKNLQENISEWK 1065
Query 176 Q 176
+
Sbjct 1066 K 1066
> tgo:TGME49_057080 cAMP-specific 3',5'-cyclic phosphodiesterase,
putative (EC:3.1.4.17)
Length=1691
Score = 115 bits (287), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 57/164 (34%), Positives = 95/164 (57%), Gaps = 5/164 (3%)
Query 17 NIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHFDHRKQRDDTWLIME 76
NIF+ LAK+ +R N+I LIL+TDM H ++RLR ++ +FD + D WL+ +
Sbjct 1435 NIFSGLAKEVYQYMRQNIIGLILSTDMSKHISYVSRLRVRAESGNFDVNNE-GDRWLLFQ 1493
Query 77 MCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPLCDRDLHSSMARNQV 136
C++AAD+ H+ WD H W+ + EF+ QGDEE R G + + DR ++Q
Sbjct 1494 GCIKAADLAHTATFWDNHKRWAECLCEEFFKQGDEERRQGMNVMDIFDRRQKDRFPQSQY 1553
Query 137 GFLEHLVKPLFVELHSVDSM----KGAFQEAVNTIDANISRWQQ 176
F+E +V+PLF + S++ + G + T+ +N++RW++
Sbjct 1554 RFIELVVEPLFNSVCSIEDLLKGRGGVRGKICKTLSSNLARWKE 1597
> tpv:TP01_1200 hypothetical protein
Length=840
Score = 113 bits (283), Expect = 6e-25, Method: Composition-based stats.
Identities = 60/151 (39%), Positives = 87/151 (57%), Gaps = 5/151 (3%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H LTF VL + N+F+ L K+E+ VR +I LIL+TD++ HF I + R R + F
Sbjct 680 HCCLTFNVLRQS--NLFSFLDKEEMKIVRRKIIQLILSTDLEDHFQTITQFRVRRASREF 737
Query 63 DHRKQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPL 122
+ D I +M ++A+DIG S + W EWS R+ EFY QG EE G I+PL
Sbjct 738 SVEGEIDT---ICKMLIKASDIGSSCMEWRLTLEWSQRLVQEFYTQGLEELSLGLPITPL 794
Query 123 CDRDLHSSMARNQVGFLEHLVKPLFVELHSV 153
CD + H+ + + Q GFL V PL+ E+ ++
Sbjct 795 CDPNKHNHLPKAQSGFLAIFVIPLYTEIANI 825
> pfa:MAL13P1.119 calcium/calmodulin-dependent 3',5'-cyclic nucleotide
phosphodiesterase 1b (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=769
Score = 106 bits (265), Expect = 8e-23, Method: Composition-based stats.
Identities = 55/178 (30%), Positives = 98/178 (55%), Gaps = 8/178 (4%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAIN--RLRAARQNP 60
H F +L NIF + +L +R +I LILATDM H + R+++ +
Sbjct 561 HCSYLFNILLKDENNIFKNEDPKCLLALRQQIIELILATDMSKHIKILAQFRIKSIKIKS 620
Query 61 HFDHRKQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGIS 120
+ + + L ++M ++AAD+ H+ + W +H++W R+ EFY +GDE + G I+
Sbjct 621 YIE-----KNIILCLKMIIKAADLSHNCVDWSEHYQWVKRLVNEFYYEGDELFQMGYKIN 675
Query 121 PLCDRDLHSSMARNQVGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDANISRWQQLQ 178
PL DR+ H++ + Q FL+ LV PL + L ++D+ Q+ +N + N S+W +++
Sbjct 676 PLFDRNCHNNFIQIQRTFLKELVYPLIISLKTLDNTS-ITQDMINNVKRNYSKWTKIE 732
> mmu:18573 Pde1a, AI987702, AW125737, MGC116577; phosphodiesterase
1A, calmodulin-dependent (EC:3.1.4.17); K13755 calcium/calmodulin-dependent
3',5'-cyclic nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=545
Score = 105 bits (262), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 50/140 (35%), Positives = 83/140 (59%), Gaps = 7/140 (5%)
Query 8 FKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHFDHRKQ 67
++++ NI +L+KD+ ++R+ +I ++LATDM HF I +R + Q P R +
Sbjct 297 YRLMQEEEMNILVNLSKDDWRDLRNLVIEMVLATDMSGHFQQIKNIRNSLQQPEGIDRAK 356
Query 68 RDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPLCDRDL 127
M + + AADI H +W H+ W+ + EF+LQGD+EA G SPLCDR
Sbjct 357 ------TMSLILHAADISHPAKTWKLHYRWTMALMEEFFLQGDKEAELGLPFSPLCDRK- 409
Query 128 HSSMARNQVGFLEHLVKPLF 147
+ +A++Q+GF++ +V+P F
Sbjct 410 STMVAQSQIGFIDFIVEPTF 429
> hsa:5136 PDE1A, HCAM1, HSPDE1A, MGC26303; phosphodiesterase
1A, calmodulin-dependent (EC:3.1.4.17); K13755 calcium/calmodulin-dependent
3',5'-cyclic nucleotide phosphodiesterase [EC:3.1.4.17]
Length=535
Score = 105 bits (261), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 50/140 (35%), Positives = 83/140 (59%), Gaps = 7/140 (5%)
Query 8 FKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHFDHRKQ 67
++++ NI +L+KD+ ++R+ +I ++L+TDM HF I +R + Q P R +
Sbjct 297 YRLMQEEEMNILINLSKDDWRDLRNLVIEMVLSTDMSGHFQQIKNIRNSLQQPEGIDRAK 356
Query 68 RDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPLCDRDL 127
M + + AADI H SW H+ W+ + EF+LQGD+EA G SPLCDR
Sbjct 357 ------TMSLILHAADISHPAKSWKLHYRWTMALMEEFFLQGDKEAELGLPFSPLCDRK- 409
Query 128 HSSMARNQVGFLEHLVKPLF 147
+ +A++Q+GF++ +V+P F
Sbjct 410 STMVAQSQIGFIDFIVEPTF 429
> hsa:5153 PDE1B, PDE1B1, PDES1B; phosphodiesterase 1B, calmodulin-dependent
(EC:3.1.4.17); K13755 calcium/calmodulin-dependent
3',5'-cyclic nucleotide phosphodiesterase [EC:3.1.4.17]
Length=536
Score = 103 bits (256), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 53/146 (36%), Positives = 84/146 (57%), Gaps = 7/146 (4%)
Query 8 FKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHFDHRKQ 67
F+++ NIF +L KDE +E+R+ +I ++LATDM HF + ++ A Q + +
Sbjct 301 FRLMQDDEMNIFINLTKDEFVELRALVIEMVLATDMSCHFQQVKTMKTALQ------QLE 354
Query 68 RDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPLCDRDL 127
R D + + + AADI H W H W+ + EF+ QGD+EA G SPLCDR
Sbjct 355 RIDKPKALSLLLHAADISHPTKQWLVHSRWTKALMEEFFRQGDKEAELGLPFSPLCDRT- 413
Query 128 HSSMARNQVGFLEHLVKPLFVELHSV 153
+ +A++Q+GF++ +V+P F L V
Sbjct 414 STLVAQSQIGFIDFIVEPTFSVLTDV 439
> mmu:18574 Pde1b, 63kDa, Pde1b1; phosphodiesterase 1B, Ca2+-calmodulin
dependent (EC:3.1.4.17); K13755 calcium/calmodulin-dependent
3',5'-cyclic nucleotide phosphodiesterase [EC:3.1.4.17]
Length=535
Score = 103 bits (256), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 53/146 (36%), Positives = 83/146 (56%), Gaps = 7/146 (4%)
Query 8 FKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHFDHRKQ 67
F+++ NIF +L KDE E+R+ +I ++LATDM HF + ++ A Q + +
Sbjct 300 FRMMQDDEMNIFINLTKDEFAELRALVIEMVLATDMSCHFQQVKTMKTALQ------QLE 353
Query 68 RDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPLCDRDL 127
R D + + + AADI H W H W+ + EF+ QGD+EA G SPLCDR
Sbjct 354 RIDKSKALSLLLHAADISHPTKQWSVHSRWTKALMEEFFRQGDKEAELGLPFSPLCDRT- 412
Query 128 HSSMARNQVGFLEHLVKPLFVELHSV 153
+ +A++Q+GF++ +V+P F L V
Sbjct 413 STLVAQSQIGFIDFIVEPTFSVLTDV 438
> dre:564465 pde1a; phosphodiesterase 1A, calmodulin-dependent;
K13755 calcium/calmodulin-dependent 3',5'-cyclic nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=625
Score = 98.6 bits (244), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 48/145 (33%), Positives = 82/145 (56%), Gaps = 7/145 (4%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H ++++ NI +L+KD+ E+R+ +I ++++TDM HF I +R + Q P
Sbjct 295 HVSAAYRLMQEDEMNILVNLSKDDWRELRTLVIEMVMSTDMSCHFQQIKTMRNSLQQP-- 352
Query 63 DHRKQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPL 122
+ D + + + AADI H W H+ W+ + EF+ QGD+EA G SPL
Sbjct 353 ----EGIDKAKALSLMLHAADISHPAKGWKLHYRWTQALMEEFFRQGDKEAELGLPFSPL 408
Query 123 CDRDLHSSMARNQVGFLEHLVKPLF 147
CDR + +A++Q+GF++ +V+P F
Sbjct 409 CDRKA-TMIAQSQIGFIDFIVEPTF 432
> bbo:BBOV_IV010070 23.m06154; 3'5'-cyclic nucleotide phosphodiesterase
(EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=485
Score = 97.4 bits (241), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 51/172 (29%), Positives = 89/172 (51%), Gaps = 6/172 (3%)
Query 30 VRSNMISLILATDMKSHFGAINRLRAARQNPHFDHRKQRDDTWLIMEMCMRAADIGHSIL 89
+R +I L+LATDM +HF +N ++ R + FD+ +D W + + ++ ADIGH+ L
Sbjct 310 IRKRIIQLVLATDMMTHFTHVNNVKERRLSGTFDYVNNPEDLWFCLVLLVKTADIGHNFL 369
Query 90 SWDQHFEWSSRVTAEFYLQGDEEARHGRGISPLCDRDLHSSMARNQVGFLEHLVKPLFVE 149
W +H W++ + EF++QGDEE + DR + + +Q+GF + PL E
Sbjct 370 PWCEHLPWTNALFDEFHMQGDEERLLSLPLLLFFDRTRSADIPDSQLGFFQGFTSPLIDE 429
Query 150 LHSVDSMKGAFQEAVN-TIDANISRWQ-----QLQDIGVDVTFSPVVMACED 195
L +++ + +N N+ W+ QL +I +D+ S V +D
Sbjct 430 LTFINTNSTYLNDFLNANAHDNLEHWKKNSKRQLVEIVIDLEKSEEVFEIKD 481
> mmu:18575 Pde1c; phosphodiesterase 1C (EC:3.1.4.17); K13755
calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=631
Score = 96.7 bits (239), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 49/135 (36%), Positives = 76/135 (56%), Gaps = 7/135 (5%)
Query 16 CNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHFDHRKQRDDTWLIM 75
NI +L+KD+ E R+ +I +++ATDM HF I ++ A Q P + + +
Sbjct 315 MNILVNLSKDDWREFRTLVIEMVMATDMSCHFQQIKAMKTALQQPEAIEKPK------AL 368
Query 76 EMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPLCDRDLHSSMARNQ 135
+ + ADI H +WD H W+ + EF+ QGD EA G SPLCDR + +A++Q
Sbjct 369 SLMLHTADISHPAKAWDLHHRWTMSLLEEFFRQGDREAELGLPFSPLCDRK-STMVAQSQ 427
Query 136 VGFLEHLVKPLFVEL 150
VGF++ +V+P F L
Sbjct 428 VGFIDFIVEPTFTVL 442
> hsa:5137 PDE1C, Hcam3; phosphodiesterase 1C, calmodulin-dependent
70kDa (EC:3.1.4.17); K13755 calcium/calmodulin-dependent
3',5'-cyclic nucleotide phosphodiesterase [EC:3.1.4.17]
Length=634
Score = 96.3 bits (238), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 49/135 (36%), Positives = 76/135 (56%), Gaps = 7/135 (5%)
Query 16 CNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHFDHRKQRDDTWLIM 75
NI +L+KD+ E R+ +I +++ATDM HF I ++ A Q P + + +
Sbjct 315 MNILINLSKDDWREFRTLVIEMVMATDMSCHFQQIKAMKTALQQPEAIEKPK------AL 368
Query 76 EMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPLCDRDLHSSMARNQ 135
+ + ADI H +WD H W+ + EF+ QGD EA G SPLCDR + +A++Q
Sbjct 369 SLMLHTADISHPAKAWDLHHRWTMSLLEEFFRQGDREAELGLPFSPLCDRK-STMVAQSQ 427
Query 136 VGFLEHLVKPLFVEL 150
VGF++ +V+P F L
Sbjct 428 VGFIDFIVEPTFTVL 442
> tpv:TP01_0664 3',5' cyclic nucleotide phosphodiesterase; K01120
3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=450
Score = 95.1 bits (235), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 57/157 (36%), Positives = 86/157 (54%), Gaps = 12/157 (7%)
Query 6 LTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHFDHR 65
L F ++ + + N ++ + KD +R +I LIL+TDM +HF I+ ++ R FD
Sbjct 246 LLFYIIKNNT-NFYSIIPKDIWPSLRKRIIQLILSTDMINHFLHIDNVKEKRLQGKFD-I 303
Query 66 KQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPLC-- 123
K DD WL++ +C++AADIGH+ L W H W+ + EFY QGDEE + ISPL
Sbjct 304 KNDDDFWLLLVLCIKAADIGHNFLPWADHLPWTQVLFEEFYKQGDEEKM--QSISPLMFF 361
Query 124 DRDLHSSMARNQVGFLEH------LVKPLFVELHSVD 154
DR+ ++ Q+ F L +PL EL+ D
Sbjct 362 DRNESRNIPILQLQFFNTMTQLLILSQPLLSELNYFD 398
> bbo:BBOV_IV006510 23.m05980; 3'5'-cyclic nucleotide phosphodiesterase
(EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=777
Score = 91.7 bits (226), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 59/184 (32%), Positives = 87/184 (47%), Gaps = 10/184 (5%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINR--LRAARQNP 60
H +VL CN+F D VRS +I ILATD F I++ L R +
Sbjct 571 HAASALRVLCTPGCNLFPDCDYDY---VRSQIIEHILATDTVDQFHMISQFVLSCKRDDF 627
Query 61 HFDHRKQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGIS 120
F+ R L +M M+AAD+ + W EW SR+ EFY QG EE + IS
Sbjct 628 TFEEYNSRI---LTSKMLMKAADVAAPTMQWANSLEWVSRLLGEFYAQGAEEVDYCIPIS 684
Query 121 PLCDRDLHSSMARNQVGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDANISRWQQLQDI 180
LCDR H A++Q FL +V PL+ + S+ S + ++ ++ N +W ++
Sbjct 685 ALCDRQHHEQSAKSQAAFLRIVVAPLYEAIASLGSPE--IDAIMDQLEENAEKWNEMDKA 742
Query 181 GVDV 184
G +
Sbjct 743 GTII 746
> mmu:18583 Pde7a, AU015378, AW047537; phosphodiesterase 7A (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=482
Score = 87.8 bits (216), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 52/164 (31%), Positives = 93/164 (56%), Gaps = 7/164 (4%)
Query 17 NIFASLAKDEILEVRSNMISLILATDM--KSHFGAINRLRAARQNPHFDHRKQRDDTWLI 74
+F+ L + E+ + + +LILATD+ ++ + ++ R + + H D + R L+
Sbjct 297 GLFSHLPLESRQEMEAQIGALILATDISRQNEYLSLFRSHLDKGDLHLDDGRHRH---LV 353
Query 75 MEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPLCDRDLHSSMARN 134
++M ++ ADI + +W+ +WS +VT EF+ QGD E ++ G+SPLCDR S+A
Sbjct 354 LQMALKCADICNPCRNWELSKQWSEKVTEEFFHQGDIEKKYHLGVSPLCDRQT-ESIANI 412
Query 135 QVGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDANISRWQQLQ 178
Q+GF+ +LV+PLF E + + Q + + N + W+ LQ
Sbjct 413 QIGFMTYLVEPLFTEWARFSDTRLS-QTMLGHVGLNKASWKGLQ 455
> cel:T04D3.3 pde-1; PhosphoDiEsterase family member (pde-1);
K13755 calcium/calmodulin-dependent 3',5'-cyclic nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=664
Score = 86.7 bits (213), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 50/143 (34%), Positives = 77/143 (53%), Gaps = 11/143 (7%)
Query 8 FKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPH-FDHRK 66
F+++ NI L +DE E+R+ +I ++LATDM +HF I +++ P D K
Sbjct 411 FRLMKEDDKNILTHLTRDEYKELRNMVIEIVLATDMSTHFMQIKTMKSMLSLPEGIDKNK 470
Query 67 QRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPLCDR- 125
+ + + A DI H W+ H W+ V EF+ QGD EA G SPLCDR
Sbjct 471 A-------LCLIVHACDISHPAKPWNLHERWTEGVLEEFFRQGDLEASMGLPYSPLCDRH 523
Query 126 DLHSSMARNQVGFLEHLVKPLFV 148
+H +A +Q+GF++ +V+P V
Sbjct 524 TVH--VADSQIGFIDFIVEPTMV 544
> mmu:18585 Pde9a, PDE9A1; phosphodiesterase 9A (EC:3.1.4.35);
K13761 high affinity cGMP-specific 3',5'-cyclic phosphodiesterase
9 [EC:3.1.4.35]
Length=534
Score = 84.3 bits (207), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 44/145 (30%), Positives = 78/145 (53%), Gaps = 4/145 (2%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H + F++L+ CNIFAS+ + ++R MI+LILATDM H ++ + +N F
Sbjct 324 HCAIAFQILARPECNIFASVPPEGFRQIRQGMITLILATDMARHAEIMDSFKEKMEN--F 381
Query 63 DHRKQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPL 122
D+ + T L M + ++ DI + + + W + E+++Q D E G ++P
Sbjct 382 DYSNEEHLTLLKM-ILIKCCDISNEVRPMEVAEPWVDCLLEEYFMQSDREKSEGLPVAPF 440
Query 123 CDRDLHSSMARNQVGFLEHLVKPLF 147
DRD + A Q+GF++ ++ P+F
Sbjct 441 MDRD-KVTKATAQIGFIKFVLIPMF 464
> hsa:5152 PDE9A, FLJ90181, HSPDE9A2; phosphodiesterase 9A (EC:3.1.4.35);
K13761 high affinity cGMP-specific 3',5'-cyclic
phosphodiesterase 9 [EC:3.1.4.35]
Length=533
Score = 84.0 bits (206), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 43/145 (29%), Positives = 78/145 (53%), Gaps = 4/145 (2%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H + F++L+ CNIF+++ D ++R MI+LILATDM H ++ + +N F
Sbjct 325 HCAVAFQILAEPECNIFSNIPPDGFKQIRQGMITLILATDMARHAEIMDSFKEKMEN--F 382
Query 63 DHRKQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPL 122
D+ + T L M + ++ DI + + + W + E+++Q D E G ++P
Sbjct 383 DYSNEEHMTLLKM-ILIKCCDISNEVRPMEVAEPWVDCLLEEYFMQSDREKSEGLPVAPF 441
Query 123 CDRDLHSSMARNQVGFLEHLVKPLF 147
DRD + A Q+GF++ ++ P+F
Sbjct 442 MDRD-KVTKATAQIGFIKFVLIPMF 465
> mmu:18578 Pde4b, Dpde4, R74983, dunce; phosphodiesterase 4B,
cAMP specific (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=564
Score = 83.2 bits (204), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 49/180 (27%), Positives = 94/180 (52%), Gaps = 8/180 (4%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H + FK+L C+IF +L K + +R +I ++LATDM H + L+ +
Sbjct 307 HLAVGFKLLQEEHCDIFQNLTKKQRQTLRKMVIDMVLATDMSKHMSLLADLKTMVETKKV 366
Query 63 DHR-----KQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGR 117
D ++ + AD+ + S + + +W+ R+ EF+ QGD+E G
Sbjct 367 TSSGVLLLDNYTDRIQVLRNMVHCADLSNPTKSLELYRQWTDRIMEEFFQQGDKERERGM 426
Query 118 GISPLCDRDLHSSMARNQVGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDANISRWQQL 177
ISP+CD+ +S+ ++QVGF++++V PL+ D ++ Q+ ++T++ N + +Q +
Sbjct 427 EISPMCDKHT-ASVEKSQVGFIDYIVHPLWETW--ADLVQPDAQDILDTLEDNRNWYQSM 483
> hsa:5142 PDE4B, DKFZp686F2182, DPDE4, MGC126529, PDE4B5, PDEIVB;
phosphodiesterase 4B, cAMP-specific (EC:3.1.4.17); K01120
3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=564
Score = 82.8 bits (203), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 49/180 (27%), Positives = 94/180 (52%), Gaps = 8/180 (4%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H + FK+L C+IF +L K + +R +I ++LATDM H + L+ +
Sbjct 307 HLAVGFKLLQEEHCDIFMNLTKKQRQTLRKMVIDMVLATDMSKHMSLLADLKTMVETKKV 366
Query 63 DHR-----KQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGR 117
D ++ + AD+ + S + + +W+ R+ EF+ QGD+E G
Sbjct 367 TSSGVLLLDNYTDRIQVLRNMVHCADLSNPTKSLELYRQWTDRIMEEFFQQGDKERERGM 426
Query 118 GISPLCDRDLHSSMARNQVGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDANISRWQQL 177
ISP+CD+ +S+ ++QVGF++++V PL+ D ++ Q+ ++T++ N + +Q +
Sbjct 427 EISPMCDKHT-ASVEKSQVGFIDYIVHPLWETW--ADLVQPDAQDILDTLEDNRNWYQSM 483
> hsa:5150 PDE7A, HCP1, PDE7; phosphodiesterase 7A (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=456
Score = 82.8 bits (203), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 50/163 (30%), Positives = 93/163 (57%), Gaps = 7/163 (4%)
Query 18 IFASLAKDEILEVRSNMISLILATDM--KSHFGAINRLRAARQNPHFDHRKQRDDTWLIM 75
+F+ L + ++ + + +LILATD+ ++ + ++ R R + + + R L++
Sbjct 272 LFSHLPLESRQQMETQIGALILATDISRQNEYLSLFRSHLDRGDLCLEDTRHRH---LVL 328
Query 76 EMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPLCDRDLHSSMARNQ 135
+M ++ ADI + +W+ +WS +VT EF+ QGD E ++ G+SPLCDR S+A Q
Sbjct 329 QMALKCADICNPCRTWELSKQWSEKVTEEFFHQGDIEKKYHLGVSPLCDRHT-ESIANIQ 387
Query 136 VGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDANISRWQQLQ 178
+GF+ +LV+PLF E + + + Q + + N + W+ LQ
Sbjct 388 IGFMTYLVEPLFTEWARFSNTRLS-QTMLGHVGLNKASWKGLQ 429
> dre:565259 pde4c, im:7160317, si:dkey-149i17.5; phosphodiesterase
4C, cAMP-specific (phosphodiesterase E1 dunce homolog,
Drosophila)
Length=672
Score = 82.8 bits (203), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 49/180 (27%), Positives = 93/180 (51%), Gaps = 8/180 (4%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H + FK+L +C+IF +L K + +R +I ++LATDM H + L+ +
Sbjct 447 HLAVGFKLLQEENCDIFQNLTKKQRQSLRKMVIDMVLATDMSKHMNLLADLKTMVETKKV 506
Query 63 DHR-----KQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGR 117
D +++ + AD+ + + + +W+ R+ EF+ QGD E G
Sbjct 507 TSLGVLLLDNYSDRIQVLQNMVHCADLSNPTKPLELYRQWTDRIMVEFFTQGDRERDKGM 566
Query 118 GISPLCDRDLHSSMARNQVGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDANISRWQQL 177
ISP+CD+ ++S+ ++QVGF++++V PL+ D + QE ++T++ N +Q +
Sbjct 567 EISPMCDKH-NASIEKSQVGFIDYIVHPLWETW--ADLVHPDAQEILDTLEDNREWYQSM 623
> xla:444562 pde4b, MGC83972; phosphodiesterase 4B, cAMP-specific;
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=721
Score = 82.8 bits (203), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 50/180 (27%), Positives = 93/180 (51%), Gaps = 8/180 (4%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H + FK+L C+IF +L K + +R +I L+LATDM H + L+ +
Sbjct 465 HLAVGFKLLQEEHCDIFQNLTKKQRQSLRKMVIDLVLATDMSKHMSLLADLKTMVETKKV 524
Query 63 DHR-----KQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGR 117
D ++ + AD+ + S + + +W+ R+ EF+ QGD E G
Sbjct 525 TSSGVLLLDNYTDRIQVLRNMVHCADLSNPTKSLELYRQWTDRIMEEFFQQGDRERERGM 584
Query 118 GISPLCDRDLHSSMARNQVGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDANISRWQQL 177
ISP+CD+ +S+ ++QVGF++++V PL+ D ++ Q+ ++T++ N + +Q +
Sbjct 585 EISPMCDKHT-ASVEKSQVGFIDYIVHPLWETWG--DLVQPDAQDILDTLEDNRNWYQSM 641
> mmu:238871 Pde4d, 9630011N22Rik, Dpde3; phosphodiesterase 4D,
cAMP specific (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=747
Score = 82.4 bits (202), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 55/217 (25%), Positives = 103/217 (47%), Gaps = 12/217 (5%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H + FK+L +C+IF +L K + +R +I ++LATDM H + L+ +
Sbjct 474 HLAVGFKLLQEENCDIFQNLTKKQRQSLRKMVIDIVLATDMSKHMNLLADLKTMVETKKV 533
Query 63 DHR-----KQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGR 117
D +++ + AD+ + + +W+ R+ EF+ QGD E G
Sbjct 534 TSSGVLLLDNYSDRIQVLQNMVHCADLSNPTKPLQLYRQWTDRIMEEFFRQGDRERERGM 593
Query 118 GISPLCDRDLHSSMARNQVGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDANISRWQQL 177
ISP+CD+ ++S+ ++QVGF++++V PL+ D + Q+ ++T++ N +Q
Sbjct 594 EISPMCDKH-NASVEKSQVGFIDYIVHPLWETW--ADLVHPDAQDILDTLEDNREWYQS- 649
Query 178 QDIGVDVTFSPVVMACEDRLKGKDHVIDVSLITETRC 214
+ + SP E+ +G+ L E C
Sbjct 650 ---TIPQSPSPAPDDQEEGRQGQTEKFQFELTLEEDC 683
> dre:565706 cAMP-specific 3,5-cyclic phosphodiesterase 4B-like;
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=713
Score = 82.0 bits (201), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 49/180 (27%), Positives = 93/180 (51%), Gaps = 8/180 (4%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H + FK+L +C+IF +L K + +R +I ++LATDM H + L+ +
Sbjct 457 HLAVGFKLLQEDNCDIFQNLTKKQRTSLRRMVIDMVLATDMSKHMSLLADLKTMVETKKV 516
Query 63 DHR-----KQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGR 117
D ++ + AD+ + S + + +W+ R+ EF+ QGD E G
Sbjct 517 TSSGVLLLDNYTDRIQVLRNMVHCADLSNPTKSLELYRQWTDRIMEEFFHQGDRERERGM 576
Query 118 GISPLCDRDLHSSMARNQVGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDANISRWQQL 177
ISP+CD+ +S+ ++QVGF++++V PL+ D + Q+ ++T++ N + +Q +
Sbjct 577 EISPMCDKHT-ASVEKSQVGFIDYIVHPLWETW--ADLVHPDAQDILDTLEDNRNWYQSM 633
> cel:R153.1 pde-4; PhosphoDiEsterase family member (pde-4); K01120
3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=673
Score = 82.0 bits (201), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 41/150 (27%), Positives = 84/150 (56%), Gaps = 6/150 (4%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H + FK+L ++C+ A+L++ + L+ R +I ++LATDM H + L+ +
Sbjct 479 HLAVAFKLLQDSNCDFLANLSRKQRLQFRKIVIDMVLATDMSKHMSLLADLKTMVEAKKV 538
Query 63 DHRK-----QRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGR 117
+ +D +++ + AD+ + + + +W+ R+ E++ QGD+E G
Sbjct 539 AGNNVIVLDKYNDKIQVLQSMIHLADLSNPTKPIELYQQWNQRIMEEYWRQGDKEKELGL 598
Query 118 GISPLCDRDLHSSMARNQVGFLEHLVKPLF 147
ISP+CDR + ++ ++QVGF++++V PL+
Sbjct 599 EISPMCDRG-NVTIEKSQVGFIDYIVHPLY 627
> mmu:18577 Pde4a, D9Ertd60e, Dpde2; phosphodiesterase 4A, cAMP
specific (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=610
Score = 81.6 bits (200), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 44/150 (29%), Positives = 80/150 (53%), Gaps = 6/150 (4%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H + FK+L +C+IF +L+K + +R +I ++LATDM H + L+ +
Sbjct 258 HLAVGFKLLQEENCDIFQNLSKRQRQSLRKMVIDMVLATDMSKHMTLLADLKTMVETKKV 317
Query 63 DHR-----KQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGR 117
D ++ + AD+ + + + +W+ R+ AEF+ QGD E G
Sbjct 318 TSSGVLLLDNYSDRIQVLRNMVHCADLSNPTKPLELYRQWTDRIMAEFFQQGDRERERGM 377
Query 118 GISPLCDRDLHSSMARNQVGFLEHLVKPLF 147
ISP+CD+ +S+ ++QVGF++++V PL+
Sbjct 378 EISPMCDKHT-ASVEKSQVGFIDYIVHPLW 406
> hsa:5141 PDE4A, DPDE2, PDE4, PDE46; phosphodiesterase 4A, cAMP-specific
(EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=886
Score = 81.6 bits (200), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 49/173 (28%), Positives = 90/173 (52%), Gaps = 8/173 (4%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H + FK+L +C+IF +L+K + +R +I ++LATDM H + L+ +
Sbjct 506 HLAVGFKLLQEDNCDIFQNLSKRQRQSLRKMVIDMVLATDMSKHMTLLADLKTMVETKKV 565
Query 63 DHR-----KQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGR 117
D ++ + AD+ + + + +W+ R+ AEF+ QGD E G
Sbjct 566 TSSGVLLLDNYSDRIQVLRNMVHCADLSNPTKPLELYRQWTDRIMAEFFQQGDRERERGM 625
Query 118 GISPLCDRDLHSSMARNQVGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDAN 170
ISP+CD+ +S+ ++QVGF++++V PL+ D + QE ++T++ N
Sbjct 626 EISPMCDKHT-ASVEKSQVGFIDYIVHPLWETW--ADLVHPDAQEILDTLEDN 675
> dre:566998 novel protein similar to vertebrate phosphodiesterase
4D, cAMP-specific (phosphodiesterase E3 dunce homolog,
Drosophila) (PDE4D); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=745
Score = 81.6 bits (200), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 43/150 (28%), Positives = 79/150 (52%), Gaps = 6/150 (4%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H + FK+L +C+IF +L K + +R +I ++LATDM H + L+ +
Sbjct 447 HLAVGFKLLQEENCDIFQNLTKKQRQSLRKMVIDIVLATDMSKHMNLLADLKTMVETKKV 506
Query 63 DHR-----KQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGR 117
D +++ + AD+ + + +W+ R+ EF+ QGD E G
Sbjct 507 TSSGVLLLDNYSDRIQVLQNMVHCADLSNPTKPLQLYRQWTDRIMEEFFSQGDRERERGM 566
Query 118 GISPLCDRDLHSSMARNQVGFLEHLVKPLF 147
ISP+CD+ ++S+ ++QVGF++++V PL+
Sbjct 567 EISPMCDKH-NASVEKSQVGFIDYIVHPLW 595
> tgo:TGME49_057950 3'5'-cyclic nucleotide phosphodiesterase domain-containing
protein (EC:3.1.4.17)
Length=587
Score = 80.9 bits (198), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 43/111 (38%), Positives = 63/111 (56%), Gaps = 1/111 (0%)
Query 31 RSNMISLILATDMKSHFGAINRLRAARQNPHFDHRKQRDDTWLIMEMCMRAADIGHSILS 90
R +I LI+ TDM HF + + RQ D +D ++++M ++AADIGH+ +
Sbjct 286 RKEVIELIVHTDMTKHFSLLALFKVKRQTGGLDIVNNEEDRSMLLKMLLKAADIGHATKA 345
Query 91 WDQHFEWSSRVTAEFYLQGDEEARHGRGISPLCDRDLH-SSMARNQVGFLE 140
+D HF S + EF+ QGDEE + G ISPLCDR + +Q GFL+
Sbjct 346 FDFHFFLSCCLIEEFHRQGDEEKKRGMVISPLCDRQQELRQIFSSQAGFLQ 396
> hsa:5144 PDE4D, DKFZp686M11213, DPDE3, FLJ97311, HSPDE4D, PDE4DN2,
STRK1; phosphodiesterase 4D, cAMP-specific (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=809
Score = 80.9 bits (198), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 43/150 (28%), Positives = 79/150 (52%), Gaps = 6/150 (4%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H + FK+L +C+IF +L K + +R +I ++LATDM H + L+ +
Sbjct 535 HLAVGFKLLQEENCDIFQNLTKKQRQSLRKMVIDIVLATDMSKHMNLLADLKTMVETKKV 594
Query 63 DHR-----KQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGR 117
D +++ + AD+ + + +W+ R+ EF+ QGD E G
Sbjct 595 TSSGVLLLDNYSDRIQVLQNMVHCADLSNPTKPLQLYRQWTDRIMEEFFRQGDRERERGM 654
Query 118 GISPLCDRDLHSSMARNQVGFLEHLVKPLF 147
ISP+CD+ ++S+ ++QVGF++++V PL+
Sbjct 655 EISPMCDKH-NASVEKSQVGFIDYIVHPLW 683
> dre:562357 phosphodiesterase 9A-like
Length=529
Score = 80.5 bits (197), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 49/167 (29%), Positives = 80/167 (47%), Gaps = 13/167 (7%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H + F++L T NIF +L ++ +R +I ILATDM H +N+ +A F
Sbjct 324 HCAVAFEILEKTESNIFRNLPTEQYKRIREGIIKCILATDMSRHTDILNKFKAILPVFDF 383
Query 63 DHRKQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPL 122
++ RD +IM ++ +DI + D W + EF+ Q D E G ++P
Sbjct 384 GNKDHRDVLMMIM---IKVSDISNEARPMDVAEPWLDCLLQEFFNQSDMEKLEGLPVTPF 440
Query 123 CDRDLHSSMARNQVGFLEHLVKPLFVEL---------HSVDSMKGAF 160
DRD + +Q GF+ ++ PLF+EL H +D ++ A
Sbjct 441 MDRD-KVTKPSSQTGFIRFVLFPLFIELANLFPCLEQHIIDPVRKAL 486
> dre:572061 pde4a; phosphodiesterase 4A, cAMP-specific; K01120
3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=736
Score = 80.5 bits (197), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 53/181 (29%), Positives = 91/181 (50%), Gaps = 14/181 (7%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPH- 61
H + FK+L +C+IF +L K + +R +I ++LATDM H + L+ +
Sbjct 465 HLAVGFKLLHEENCDIFQNLTKRQRQSLRKLVIDMVLATDMSKHMSLLADLKTMVETKKV 524
Query 62 -------FDHRKQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEAR 114
DH R M C AD+ + + +W+ R+ EF+ QGD+E
Sbjct 525 TSSGVLMLDHYNDRIQVLRNMVHC---ADLSNPTKPLAVYRQWTERIMEEFFQQGDKERE 581
Query 115 HGRGISPLCDRDLHSSMARNQVGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDANISRW 174
G ISP+CD+ +S+ ++QVGF++++V PL+ D + QE ++T++ N +
Sbjct 582 RGMEISPMCDKHT-ASVEKSQVGFIDYIVHPLWETWG--DLVHPDAQEILDTLEDNRDWY 638
Query 175 Q 175
Q
Sbjct 639 Q 639
> dre:571344 fc68c05, si:ch211-255d18.8, wu:fc68c05; si:ch211-255d18.1
Length=709
Score = 79.7 bits (195), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 47/180 (26%), Positives = 91/180 (50%), Gaps = 8/180 (4%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H + FK+L +C+IF +L+K + +R I ++LATDM H + L+ +
Sbjct 433 HLAVGFKLLQEENCDIFCNLSKKQRQSLRQMTIDMVLATDMSKHMNFLADLKTMVETKKV 492
Query 63 DHR-----KQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGR 117
+ D +++ + AD+ + + + +W+ R+ EF+ QGD E G
Sbjct 493 TSQGVLLLDNYSDRIQVLQNMVHCADLSNPTKPLELYRKWTDRIMVEFFSQGDRERDKGI 552
Query 118 GISPLCDRDLHSSMARNQVGFLEHLVKPLFVELHSVDSMKGAFQEAVNTIDANISRWQQL 177
+SP+CD+ +SM QVGF+++++ PL+ D + Q+ ++T++ N +Q +
Sbjct 553 DVSPMCDKHT-ASMENTQVGFIDYIIHPLWETW--ADLVHPDAQDILDTLEDNREWYQSM 609
> hsa:5143 PDE4C, DPDE1, MGC126222; phosphodiesterase 4C, cAMP-specific
(EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=712
Score = 79.7 bits (195), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 43/150 (28%), Positives = 79/150 (52%), Gaps = 6/150 (4%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H + FK+L +C+IF +L+ + L +R +I ++LATDM H + L+ +
Sbjct 461 HLAVGFKLLQAENCDIFQNLSAKQRLSLRRMVIDMVLATDMSKHMNLLADLKTMVETKKV 520
Query 63 DHR-----KQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGR 117
D +++ + AD+ + + +W+ R+ AEF+ QGD E G
Sbjct 521 TSLGVLLLDNYSDRIQVLQNLVHCADLSNPTKPLPLYRQWTDRIMAEFFQQGDRERESGL 580
Query 118 GISPLCDRDLHSSMARNQVGFLEHLVKPLF 147
ISP+CD+ +S+ ++QVGF++++ PL+
Sbjct 581 DISPMCDKHT-ASVEKSQVGFIDYIAHPLW 609
> xla:495146 hypothetical LOC495146; K01120 3',5'-cyclic-nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=682
Score = 79.7 bits (195), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 43/150 (28%), Positives = 79/150 (52%), Gaps = 6/150 (4%)
Query 3 HDVLTFKVLSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHF 62
H + FK+L +C+IF +L K + +R +I ++LATDM H + L+ +
Sbjct 417 HLAVGFKLLQEENCDIFQNLPKRQRQMMRKMVIDMVLATDMSKHMSLLADLKTMVETKKV 476
Query 63 DHR-----KQRDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGR 117
D ++ + AD+ + + + +W+ R+ EF+ QGD+E G
Sbjct 477 TSSGVLLLDNYTDRIQVLRNMVHCADLSNPTKPLELYRQWTDRILEEFFRQGDKERERGM 536
Query 118 GISPLCDRDLHSSMARNQVGFLEHLVKPLF 147
ISP+CD+ +S+ ++QVGF++++V PL+
Sbjct 537 EISPMCDKHT-ASVEKSQVGFIDYIVHPLW 565
> dre:567051 si:dkey-48h7.2; K13296 cGMP-inhibited 3',5'-cyclic
phosphodiesterase [EC:3.1.4.17]
Length=1117
Score = 79.3 bits (194), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 52/151 (34%), Positives = 72/151 (47%), Gaps = 5/151 (3%)
Query 11 LSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQN---PHFDHRKQ 67
+S N ASL E R +I ILATD+K HF + A + D +
Sbjct 804 MSRPEYNFLASLDHMEFKRFRFLVIEAILATDLKKHFDFLAEFNAKVGDDGCSGIDWTNE 863
Query 68 RDDTWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPLCDRDL 127
+D L+ +MC++ ADI + D H +W+ + EFY QGDEE+ G ISP DR
Sbjct 864 -NDRLLVCQMCIKLADINGPLKCKDLHLQWTEGIVNEFYEQGDEESSLGLPISPFMDRSA 922
Query 128 HSSMARNQVGFLEHLVKPLFVELHSVDSMKG 158
+A+ Q F+ H+V PL S M G
Sbjct 923 -PQLAKLQESFITHIVGPLCSSYDSAALMPG 952
> mmu:54611 Pde3a, A930022O17Rik, C87899; phosphodiesterase 3A,
cGMP inhibited (EC:3.1.4.17); K13296 cGMP-inhibited 3',5'-cyclic
phosphodiesterase [EC:3.1.4.17]
Length=1141
Score = 77.8 bits (190), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 50/145 (34%), Positives = 69/145 (47%), Gaps = 4/145 (2%)
Query 11 LSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHFDHRKQRDD 70
+S N +L E R +I ILATD+K HF + + A + +D
Sbjct 878 MSRPEYNFLVNLDHVEFKHFRFLVIEAILATDLKKHFDFVAKFNAKVNDDVGIDWTNEND 937
Query 71 TWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPLCDRDLHSS 130
L+ +MC++ ADI + H W+ + +EFY QGDEEA G ISP DR
Sbjct 938 RLLVCQMCIKLADINGPAKCKELHLRWTEGIASEFYEQGDEEASLGLPISPFMDRSA-PQ 996
Query 131 MARNQVGFLEHLVKPLFVELHSVDS 155
+A Q F+ H+V PL HS DS
Sbjct 997 LANLQESFISHIVGPL---CHSYDS 1018
> hsa:5139 PDE3A, CGI-PDE; phosphodiesterase 3A, cGMP-inhibited
(EC:3.1.4.17); K13296 cGMP-inhibited 3',5'-cyclic phosphodiesterase
[EC:3.1.4.17]
Length=1141
Score = 77.8 bits (190), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 49/152 (32%), Positives = 69/152 (45%), Gaps = 1/152 (0%)
Query 11 LSHTSCNIFASLAKDEILEVRSNMISLILATDMKSHFGAINRLRAARQNPHFDHRKQRDD 70
+S N +L E R +I ILATD+K HF + + + +D
Sbjct 878 MSRPEYNFLINLDHVEFKHFRFLVIEAILATDLKKHFDFVAKFNGKVNDDVGIDWTNEND 937
Query 71 TWLIMEMCMRAADIGHSILSWDQHFEWSSRVTAEFYLQGDEEARHGRGISPLCDRDLHSS 130
L+ +MC++ ADI + H +W+ + EFY QGDEEA G ISP DR
Sbjct 938 RLLVCQMCIKLADINGPAKCKELHLQWTDGIVNEFYEQGDEEASLGLPISPFMDRSA-PQ 996
Query 131 MARNQVGFLEHLVKPLFVELHSVDSMKGAFQE 162
+A Q F+ H+V PL S M G + E
Sbjct 997 LANLQESFISHIVGPLCNSYDSAGLMPGKWVE 1028
Lambda K H
0.323 0.134 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8700848608
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40