bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0985_orf1
Length=240
Score E
Sequences producing significant alignments: (Bits) Value
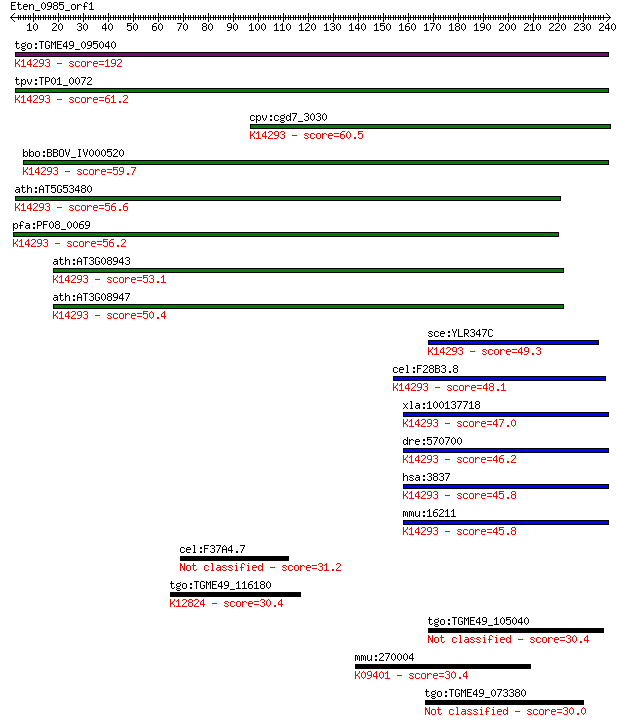
tgo:TGME49_095040 importin subunit beta-1, putative ; K14293 i... 192 9e-49
tpv:TP01_0072 importin beta; K14293 importin subunit beta-1 61.2 3e-09
cpv:cgd7_3030 importin/karyopherin ; K14293 importin subunit b... 60.5 5e-09
bbo:BBOV_IV000520 21.m02834; importin beta subunit; K14293 imp... 59.7 9e-09
ath:AT5G53480 importin beta-2, putative; K14293 importin subun... 56.6 8e-08
pfa:PF08_0069 importin beta, putative; K14293 importin subunit... 56.2 9e-08
ath:AT3G08943 binding / protein transporter; K14293 importin s... 53.1 8e-07
ath:AT3G08947 binding / protein transporter; K14293 importin s... 50.4 6e-06
sce:YLR347C KAP95, RSL1; Kap95p; K14293 importin subunit beta-1 49.3 1e-05
cel:F28B3.8 imb-1; IMportin Beta family member (imb-1); K14293... 48.1 3e-05
xla:100137718 kpnb1, imb1, impnb, ipo1, ipob, ntf97; karyopher... 47.0 6e-05
dre:570700 kpnb1, MGC123184, wu:fc12b10, zgc:123184; karyopher... 46.2 9e-05
hsa:3837 KPNB1, IMB1, IPO1, IPOB, Impnb, MGC2155, MGC2156, MGC... 45.8 1e-04
mmu:16211 Kpnb1, AA409963, IPOB, Impnb, MGC8315; karyopherin (... 45.8 1e-04
cel:F37A4.7 rbf-1; RaBPHilin family member (rbf-1) 31.2 3.9
tgo:TGME49_116180 FF domain-containing protein ; K12824 transc... 30.4 5.8
tgo:TGME49_105040 hypothetical protein 30.4 6.0
mmu:270004 Foxi2, B130055A05Rik; forkhead box I2; K09401 forkh... 30.4 6.1
tgo:TGME49_073380 hypothetical protein 30.0 7.7
> tgo:TGME49_095040 importin subunit beta-1, putative ; K14293
importin subunit beta-1
Length=971
Score = 192 bits (488), Expect = 9e-49, Method: Compositional matrix adjust.
Identities = 110/239 (46%), Positives = 154/239 (64%), Gaps = 12/239 (5%)
Query 3 LLLLLDHFEGQLRKSFSFEQTEQTKQRQGMLCGVIQILCLRLGDQVKPVAGKLWNTLKQV 62
+L LLDHF QL +SF FE E T+QRQG+LCGVIQILCLRLGDQV+PVA ++W L ++
Sbjct 603 MLKLLDHFVQQLSQSFMFEANEATRQRQGLLCGVIQILCLRLGDQVQPVASQIWACLARI 662
Query 63 FQ--RPTHGLQVSQGESSAAPNNVHGFIVSSEPSVCDALLALSALINASGPAVSPFAGEV 120
F+ P + + G+ S + +SE DALLA SAL+NASGP+ + FA ++
Sbjct 663 FRGSPPANAPPGTAGQLSCS---------ASESITNDALLATSALVNASGPSTAAFAEDI 713
Query 121 ADLLVLQMQHQHEPQEAAASAVAATASTPTNDSSSADELQAARICIELVGDLSRALGTEF 180
++ +++ ++ A A+ + SS+ DELQ RIC+ELVGD+SRALG F
Sbjct 714 VCIIASGLENSTGSAAGSSGGGAGAAAATSG-SSTTDELQTVRICVELVGDVSRALGPAF 772
Query 181 GSLSDALLHGFYQLLRAPSVDRSLKPVVIVAVGDVALSLGGAKFAPFTPWLIELLVQAG 239
S LL YQ+L+ P+V+R+LKP V+VAVGD A+++GG FAP+ + +L QAG
Sbjct 773 APYSSPLLARMYQMLQDPNVERALKPCVMVAVGDAAMTMGGEAFAPYLDSFMAILHQAG 831
> tpv:TP01_0072 importin beta; K14293 importin subunit beta-1
Length=873
Score = 61.2 bits (147), Expect = 3e-09, Method: Composition-based stats.
Identities = 63/242 (26%), Positives = 104/242 (42%), Gaps = 66/242 (27%)
Query 3 LLLLLDHF----EGQLRKSFSFEQTEQTKQRQGMLCGVIQILCLRLGDQVKPVAGKLWNT 58
L+ LLDHF + FS+EQ K + + G IQ+L R+G L
Sbjct 558 LMALLDHFVVLVSQMITSDFSYEQ----KLKIQSVLGAIQVLVSRVG-----FVKNLNLL 608
Query 59 LKQVFQRPTHGLQVSQGESSAAPNNVHGFIVSSEPSVCDALLALSALINASGPA-VSPFA 117
+ +F+ L V E +ALL LSAL+N A + PF
Sbjct 609 MNSIFEF----LSVELDE--------------------EALLTLSALVNVIEFAQILPFI 644
Query 118 GEVADLLVLQMQHQHEPQEAAASAVAATASTPTNDSSSADELQAARICIELVGDLSRALG 177
++ D+++ +Q E+ +I I L D+SR L
Sbjct 645 PKIVDVVLTGLQ---------------------------SEVGICKISIGLTSDVSRCLE 677
Query 178 TEFGSLSDALLHGFYQLLRAPSVDRSLKPVVIVAVGDVALSLGGAKFAPFTPWLIELLVQ 237
+ F + D + +L+ + D++LKP++IVA+GD+A+++GG F+ + + LL+Q
Sbjct 678 SPFSTYLDRFMTILINILQDVNGDKTLKPLIIVAIGDIAMAVGGT-FSSYVQNTMTLLLQ 736
Query 238 AG 239
A
Sbjct 737 AA 738
> cpv:cgd7_3030 importin/karyopherin ; K14293 importin subunit
beta-1
Length=882
Score = 60.5 bits (145), Expect = 5e-09, Method: Composition-based stats.
Identities = 39/144 (27%), Positives = 69/144 (47%), Gaps = 26/144 (18%)
Query 97 DALLALSALINASGPAVSPFAGEVADLLVLQMQHQHEPQEAAASAVAATASTPTNDSSSA 156
+++L+L++LI A SP+ E +++ +Q
Sbjct 634 ESILSLTSLIVAMSHDFSPYVNECISIIIPLIQ-------------------------GY 668
Query 157 DELQAARICIELVGDLSRALGTEFGSLSDALLHGFYQLLRAPSVDRSLKPVVIVAVGDVA 216
DEL + IELVGDL R++G + ++ LL VDR +KP+ I+A+GD++
Sbjct 669 DELDTCKYSIELVGDLVRSVGKGINPSLEIIIKTLCALLAKNDVDRKVKPLAIIALGDIS 728
Query 217 LSLGGAKFAPFTPWLIELLVQAGV 240
++L G F P+ +++L QA +
Sbjct 729 MNL-GEDFIPYAISVLQLFQQASI 751
> bbo:BBOV_IV000520 21.m02834; importin beta subunit; K14293 importin
subunit beta-1
Length=859
Score = 59.7 bits (143), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 58/236 (24%), Positives = 96/236 (40%), Gaps = 57/236 (24%)
Query 6 LLDHFEGQLRKSF-SFEQTEQTKQRQGMLCGVIQILCLRLGDQVKPVAGKLWNTLKQVFQ 64
+L+HFE L +S + + + + + RQ +CGVIQ+L R+ + P LW L + Q
Sbjct 544 MLEHFERVLAQSVVASDHSAEGRSRQETVCGVIQVLLTRV--EFLPNTQSLWLNLFAILQ 601
Query 65 RPTHGLQVSQGESSAAPNNVHGFIVSSEPSVCDALLALSALINASGPA-VSPFAGEVADL 123
SE DALL SAL+N G + +P+ + D+
Sbjct 602 DDL-----------------------SE----DALLTASALLNRMGNSEFTPYMQRLVDI 634
Query 124 LVLQMQHQHEPQEAAASAVAATASTPTNDSSSADELQAARICIELVGDLSRALGTEFGSL 183
++ +Q D A + CIEL D++R +
Sbjct 635 VITGLQR-------------------------TDLTNACKACIELASDMARVMDRHIQPY 669
Query 184 SDALLHGFYQLLRAPSVDRSLKPVVIVAVGDVALSLGGAKFAPFTPWLIELLVQAG 239
L+ L + + LKP ++ A+GD+A++ G+ F F + LL+QA
Sbjct 670 VPQLMELLLGTLSDINTHQKLKPAIVTALGDIAMA-SGSGFNNFVEPSMRLLLQAA 724
> ath:AT5G53480 importin beta-2, putative; K14293 importin subunit
beta-1
Length=870
Score = 56.6 bits (135), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 50/218 (22%), Positives = 92/218 (42%), Gaps = 47/218 (21%)
Query 3 LLLLLDHFEGQLRKSFSFEQTEQTKQRQGMLCGVIQILCLRLGDQVKPVAGKLWNTLKQV 62
++ L + EG+ S ++ E+ + QG+LCG +Q++ +LG + P K Q+
Sbjct 553 MMELHNTLEGE---KLSLDEREKQNELQGLLCGCLQVIIQKLGSE--PTKSKFMEYADQM 607
Query 63 FQRPTHGLQVSQGESSAAPNNVHGFIVSSEPSVCDALLALSALINASGPAVSPFAGEVAD 122
GL + F S + +A+LA+ AL A+GP + + E
Sbjct 608 M-----GL------------FLRVFGCRSATAHEEAMLAIGALAYAAGPNFAKYMPEFYK 650
Query 123 LLVLQMQHQHEPQEAAASAVAATASTPTNDSSSADELQAARICIELVGDLSRALGTEFGS 182
L + +Q+ +E Q + + +VGD+ RAL +
Sbjct 651 YLEMGLQN-------------------------FEEYQVCAVTVGVVGDVCRALEDKILP 685
Query 183 LSDALLHGFYQLLRAPSVDRSLKPVVIVAVGDVALSLG 220
D ++ + L + + RS+KP + GD+AL++G
Sbjct 686 YCDGIMTQLLKDLSSNQLHRSVKPPIFSCFGDIALAIG 723
> pfa:PF08_0069 importin beta, putative; K14293 importin subunit
beta-1
Length=877
Score = 56.2 bits (134), Expect = 9e-08, Method: Composition-based stats.
Identities = 47/219 (21%), Positives = 88/219 (40%), Gaps = 52/219 (23%)
Query 2 HLLLLLDHFEGQLRKSFSFEQTEQTKQRQGMLCGVIQILCLRLGDQVKPVAGKLWNTLKQ 61
+++ LL H L ++ TE+ K QG CG +Q + RLG+Q KP ++ ++ +
Sbjct 552 YMIELLSHMMYLLTNTYLNPLTEEVKSLQGYYCGTMQFIINRLGNQCKPFLKPIYLSIFR 611
Query 62 VFQRPTHGLQVSQGESSAAPNNVHGFIVSSEPSVC-DALLALSALINASGPAVSPFAGEV 120
+F+ T +C DALLA SA+IN
Sbjct 612 LFEIRT--------------------------DICEDALLACSAIINVMAEDFREHLKTF 645
Query 121 ADLLVLQMQHQHEPQEAAASAVAATASTPTNDSSSADELQAARICIELVGDLSRALGTEF 180
+++ ++ + E +ICIE++ D+ +E+
Sbjct 646 LNVIFKGLK-------------------------NVSETSTCKICIEMISDICIPWTSEY 680
Query 181 GSLSDALLHGFYQLLRAPSVDRSLKPVVIVAVGDVALSL 219
+A+L ++ L+ V S+K ++ +GD+A +L
Sbjct 681 EKEMEAILECLWEALKTFGVHDSIKISILTVLGDIATAL 719
> ath:AT3G08943 binding / protein transporter; K14293 importin
subunit beta-1
Length=871
Score = 53.1 bits (126), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 48/206 (23%), Positives = 89/206 (43%), Gaps = 47/206 (22%)
Query 18 FSFEQTEQTKQRQGMLCGVIQILCLRLG--DQVKPVAGKLWNTLKQVFQRPTHGLQVSQG 75
S + E+ + Q LCGV+Q++ +L D +KP+ + + + ++F R G S
Sbjct 567 ISTDDREKQAELQASLCGVLQVIIQKLSSRDDMKPIIVQNADDIMRLFLR-VFGCHSS-- 623
Query 76 ESSAAPNNVHGFIVSSEPSVCDALLALSALINASGPAVSPFAGEVADLLVLQMQHQHEPQ 135
+VH +A+LA+ AL A+G + E+ L + +Q+
Sbjct 624 -------SVHE----------EAMLAIGALAYATGTEFVKYMPELFKYLQMGLQN----- 661
Query 136 EAAASAVAATASTPTNDSSSADELQAARICIELVGDLSRALGTEFGSLSDALLHGFYQLL 195
+E Q I + ++GD+ RAL + D ++ Q L
Sbjct 662 --------------------FEEYQVCSITVGVIGDICRALDEKILPFCDQIMGLLIQNL 701
Query 196 RAPSVDRSLKPVVIVAVGDVALSLGG 221
++ ++ RS+KP + GD+AL++G
Sbjct 702 QSGALHRSVKPPIFSCFGDIALAIGA 727
> ath:AT3G08947 binding / protein transporter; K14293 importin
subunit beta-1
Length=873
Score = 50.4 bits (119), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 47/206 (22%), Positives = 88/206 (42%), Gaps = 47/206 (22%)
Query 18 FSFEQTEQTKQRQGMLCGVIQILCLRLG--DQVKPVAGKLWNTLKQVFQRPTHGLQVSQG 75
S + E+ + Q LCGV+Q++ +L + KP+ + + + ++F R G S
Sbjct 567 ISTDDREKQAEVQASLCGVLQVIIQKLSGREDTKPIIMQSADDIMRLFLR-VFGCHSS-- 623
Query 76 ESSAAPNNVHGFIVSSEPSVCDALLALSALINASGPAVSPFAGEVADLLVLQMQHQHEPQ 135
+VH +A+LA+ AL A+G + E+ L + +Q+
Sbjct 624 -------SVHE----------EAMLAIGALAYATGAEFVKYMPELFKYLQMGLQN----- 661
Query 136 EAAASAVAATASTPTNDSSSADELQAARICIELVGDLSRALGTEFGSLSDALLHGFYQLL 195
+E Q I + ++GD+ RAL + D ++ Q L
Sbjct 662 --------------------FEEYQVCSITVGVLGDICRALDEKILPFCDQIMGLLIQNL 701
Query 196 RAPSVDRSLKPVVIVAVGDVALSLGG 221
++ ++ RS+KP + GD+AL++G
Sbjct 702 QSGALHRSVKPPIFSCFGDIALAIGA 727
> sce:YLR347C KAP95, RSL1; Kap95p; K14293 importin subunit beta-1
Length=861
Score = 49.3 bits (116), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 40/68 (58%), Gaps = 4/68 (5%)
Query 168 LVGDLSRALGTEFGSLSDALLHGFYQLLRAPSVDRSLKPVVIVAVGDVALSLGGAKFAPF 227
+ D+S +L +F SDA+++ Q++ P+ R LKP V+ GD+A ++G A F
Sbjct 662 FIADISNSLEEDFRRYSDAMMNVLAQMISNPNARRELKPAVLSVFGDIASNIG----ADF 717
Query 228 TPWLIELL 235
P+L +++
Sbjct 718 IPYLNDIM 725
> cel:F28B3.8 imb-1; IMportin Beta family member (imb-1); K14293
importin subunit beta-1
Length=896
Score = 48.1 bits (113), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 28/85 (32%), Positives = 45/85 (52%), Gaps = 1/85 (1%)
Query 154 SSADELQAARICIELVGDLSRALGTEFGSLSDALLHGFYQLLRAPSVDRSLKPVVIVAVG 213
S+ DE Q + LV DLSRAL E D L+ L++P +DR++K V+I
Sbjct 669 SNTDETQVCAAAVGLVTDLSRALEAEIMPFMDELIQKLILCLQSPRLDRNVKVVIIGTFA 728
Query 214 DVALSLGGAKFAPFTPWLIELLVQA 238
D+A+++ A F + ++ +L A
Sbjct 729 DIAMAI-EAHFERYVGSVVPILNNA 752
> xla:100137718 kpnb1, imb1, impnb, ipo1, ipob, ntf97; karyopherin
(importin) beta 1; K14293 importin subunit beta-1
Length=876
Score = 47.0 bits (110), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 28/82 (34%), Positives = 43/82 (52%), Gaps = 1/82 (1%)
Query 158 ELQAARICIELVGDLSRALGTEFGSLSDALLHGFYQLLRAPSVDRSLKPVVIVAVGDVAL 217
E Q + LVGDL RAL + D ++ + L +V RS+KP ++ GDVAL
Sbjct 663 EYQVCLAAVGLVGDLCRALQSNILPFCDEMMQFLLENLGNENVHRSVKPQILSVFGDVAL 722
Query 218 SLGGAKFAPFTPWLIELLVQAG 239
++GG +F + ++ L QA
Sbjct 723 AIGG-EFKKYLDVVLNTLQQAS 743
> dre:570700 kpnb1, MGC123184, wu:fc12b10, zgc:123184; karyopherin
(importin) beta 1; K14293 importin subunit beta-1
Length=876
Score = 46.2 bits (108), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 43/82 (52%), Gaps = 1/82 (1%)
Query 158 ELQAARICIELVGDLSRALGTEFGSLSDALLHGFYQLLRAPSVDRSLKPVVIVAVGDVAL 217
E Q R + LV DL RAL D ++ + L +V RS+KP ++ GD+AL
Sbjct 663 EYQVCRAAVGLVCDLCRALMANILPYCDEIMQLLLENLGNENVHRSVKPQILSVFGDIAL 722
Query 218 SLGGAKFAPFTPWLIELLVQAG 239
++GG +F + +++ L QA
Sbjct 723 AIGG-EFKKYLDIVLDTLQQAS 743
> hsa:3837 KPNB1, IMB1, IPO1, IPOB, Impnb, MGC2155, MGC2156, MGC2157,
NTF97; karyopherin (importin) beta 1; K14293 importin
subunit beta-1
Length=876
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 43/82 (52%), Gaps = 1/82 (1%)
Query 158 ELQAARICIELVGDLSRALGTEFGSLSDALLHGFYQLLRAPSVDRSLKPVVIVAVGDVAL 217
E Q + LVGDL RAL + D ++ + L +V RS+KP ++ GD+AL
Sbjct 663 EYQVCLAAVGLVGDLCRALQSNIIPFCDEVMQLLLENLGNENVHRSVKPQILSVFGDIAL 722
Query 218 SLGGAKFAPFTPWLIELLVQAG 239
++GG +F + ++ L QA
Sbjct 723 AIGG-EFKKYLEVVLNTLQQAS 743
> mmu:16211 Kpnb1, AA409963, IPOB, Impnb, MGC8315; karyopherin
(importin) beta 1; K14293 importin subunit beta-1
Length=876
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 43/82 (52%), Gaps = 1/82 (1%)
Query 158 ELQAARICIELVGDLSRALGTEFGSLSDALLHGFYQLLRAPSVDRSLKPVVIVAVGDVAL 217
E Q + LVGDL RAL + D ++ + L +V RS+KP ++ GD+AL
Sbjct 663 EYQVCLAAVGLVGDLCRALQSNILPFCDEVMQLLLENLGNENVHRSVKPQILSVFGDIAL 722
Query 218 SLGGAKFAPFTPWLIELLVQAG 239
++GG +F + ++ L QA
Sbjct 723 AIGG-EFKKYLEVVLNTLQQAS 743
> cel:F37A4.7 rbf-1; RaBPHilin family member (rbf-1)
Length=1106
Score = 31.2 bits (69), Expect = 3.9, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 25/51 (49%), Gaps = 8/51 (15%)
Query 69 GLQVSQGESSAAPNNV-----HG---FIVSSEPSVCDALLALSALINASGP 111
GLQ++ G +S PN HG F SS PS+C L A+ L + P
Sbjct 394 GLQMNGGPTSPLPNGTRRNTGHGGIEFPSSSRPSICSVLQAIEPLDRSKSP 444
> tgo:TGME49_116180 FF domain-containing protein ; K12824 transcription
elongation regulator 1
Length=772
Score = 30.4 bits (67), Expect = 5.8, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 26/52 (50%), Gaps = 2/52 (3%)
Query 65 RPTHGLQVSQGESSAAPNNVHGFIVSSEPSVCDALLALSALINASGPAVSPF 116
RP H ++V+ G+SSAA H P +L L+A ASGP SP
Sbjct 35 RP-HWMRVADGDSSAAARADHAPPAPPRPETGPTILTLAA-PGASGPQASPL 84
> tgo:TGME49_105040 hypothetical protein
Length=1173
Score = 30.4 bits (67), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 30/70 (42%), Gaps = 11/70 (15%)
Query 168 LVGDLSRALGTEFGSLSDALLHGFYQLLRAPSVDRSLKPVVIVAVGDVALSLGGAKFAPF 227
+G + R L +F S HG SL + G +A ++G FAP
Sbjct 566 FMGAVRRLLSVDFPSFLKTHPHGC-----------SLLETAVEFAGALATAVGMEVFAPD 614
Query 228 TPWLIELLVQ 237
PWL+E LV+
Sbjct 615 APWLLERLVE 624
> mmu:270004 Foxi2, B130055A05Rik; forkhead box I2; K09401 forkhead
box protein I
Length=329
Score = 30.4 bits (67), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 29/70 (41%), Gaps = 1/70 (1%)
Query 139 ASAVAATASTPTNDSSSADELQAARICIELVGDLSRALGTEFGSLSDALLHGFYQLLRAP 198
+A+ STP + +S +A C+ AL FG+L D L H F L R P
Sbjct 229 GTALEPRGSTPQDPQTSPSPSEATTTCLSGFSTAMGALAGGFGALPDGLAHDF-SLRRPP 287
Query 199 SVDRSLKPVV 208
+ P +
Sbjct 288 PTAAAHSPQI 297
> tgo:TGME49_073380 hypothetical protein
Length=2972
Score = 30.0 bits (66), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 31/65 (47%), Gaps = 3/65 (4%)
Query 167 ELVGDLSRALGTEFGSLSDALLHGFYQLLRAPSVDRSLKPVVIVAVGDVA--LSLGGAKF 224
E GD SR L E GS + G Y+L + +VD S+ PV V V L G F
Sbjct 535 ERQGDASRDLRHE-GSSGGSAPRGCYELPHSAAVDPSVAPVPQVGRRSVTPMARLPGRAF 593
Query 225 APFTP 229
+P TP
Sbjct 594 SPGTP 598
Lambda K H
0.318 0.133 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8402513444
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40