bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0974_orf1
Length=247
Score E
Sequences producing significant alignments: (Bits) Value
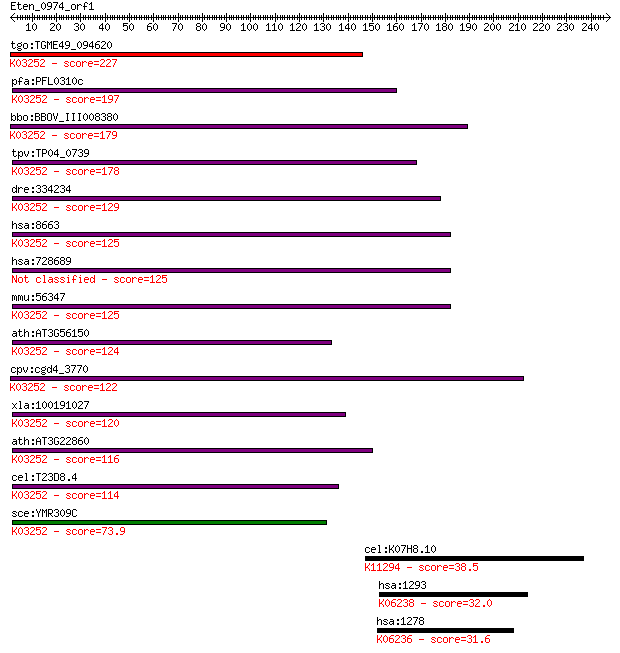
tgo:TGME49_094620 eukaryotic translation initiation factor 3 s... 227 3e-59
pfa:PFL0310c eukaryotic translation initiation factor 3 subuni... 197 2e-50
bbo:BBOV_III008380 17.m07733; eukaryotic translation initiatio... 179 6e-45
tpv:TP04_0739 eukaryotic translation initiation factor 3 subun... 178 1e-44
dre:334234 eif3c, eif3s8, fi37c04, wu:fb34a08, wu:fi37c04, zgc... 129 1e-29
hsa:8663 EIF3C, EIF3CL, EIF3S8, FLJ53378, FLJ54400, FLJ54404, ... 125 2e-28
hsa:728689 EIF3CL; eukaryotic translation initiation factor 3,... 125 2e-28
mmu:56347 Eif3c, 110kDa, 3230401O13Rik, Eif3s8, MGC36637, NIPI... 125 2e-28
ath:AT3G56150 EIF3C; EIF3C (EUKARYOTIC TRANSLATION INITIATION ... 124 4e-28
cpv:cgd4_3770 hypothetical protein ; K03252 translation initia... 122 1e-27
xla:100191027 eif3c, MGC114671, eif3-p110, eif3s8; eukaryotic ... 120 3e-27
ath:AT3G22860 TIF3C2; TIF3C2; translation initiation factor; K... 116 6e-26
cel:T23D8.4 eif-3.C; Eukaryotic Initiation Factor family membe... 114 3e-25
sce:YMR309C NIP1; Nip1p; K03252 translation initiation factor ... 73.9 5e-13
cel:K07H8.10 hypothetical protein; K11294 nucleolin 38.5 0.025
hsa:1293 COL6A3, DKFZp686D23123, DKFZp686K04147, DKFZp686N0262... 32.0 2.2
hsa:1278 COL1A2, OI4; collagen, type I, alpha 2; K06236 collag... 31.6 3.0
> tgo:TGME49_094620 eukaryotic translation initiation factor 3
subunit 8, putative ; K03252 translation initiation factor
3 subunit C
Length=1104
Score = 227 bits (578), Expect = 3e-59, Method: Compositional matrix adjust.
Identities = 109/145 (75%), Positives = 122/145 (84%), Gaps = 0/145 (0%)
Query 1 TVMAATKALQRGDWSDCARHIWSLSIWDKMFNKEHIQQMLLQKIKAEAMRTYIFTYLSLY 60
TVMAATKALQ+GDW +C R I+SL IW+K+ N IQ+ML K+K EAMRTYIFTYL+LY
Sbjct 734 TVMAATKALQKGDWKECCRFIFSLGIWEKLMNATEIQEMLKLKVKQEAMRTYIFTYLTLY 793
Query 61 DSFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASWDETSQFILISRVKHTRLQQLALTL 120
DSF+I QL MF+L + VHSIVSKMMINEEIHASWDE+SQFILISRV+ TRLQQLA TL
Sbjct 794 DSFSISQLCGMFELPESTVHSIVSKMMINEEIHASWDESSQFILISRVERTRLQQLASTL 853
Query 121 AENVANAVEQNELTLNMKNPKFALT 145
ENV NAVEQNELTLNMKNPK A T
Sbjct 854 TENVNNAVEQNELTLNMKNPKCAAT 878
> pfa:PFL0310c eukaryotic translation initiation factor 3 subunit
8, putative; K03252 translation initiation factor 3 subunit
C
Length=984
Score = 197 bits (502), Expect = 2e-50, Method: Composition-based stats.
Identities = 93/158 (58%), Positives = 117/158 (74%), Gaps = 0/158 (0%)
Query 2 VMAATKALQRGDWSDCARHIWSLSIWDKMFNKEHIQQMLLQKIKAEAMRTYIFTYLSLYD 61
++ ATK LQ+G+W C I+SLSIW K +KE +Q +L +KIK EAMRTYIF Y+S+Y+
Sbjct 790 IILATKHLQKGNWKLCCEKIFSLSIWPKFPDKEKVQNILKEKIKQEAMRTYIFRYISIYE 849
Query 62 SFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASWDETSQFILISRVKHTRLQQLALTLA 121
SF+I QL MFDL+ N VHSI+SKMMIN+EI A W+E+S+FILIS+V T LQ LAL LA
Sbjct 850 SFSIDQLCVMFDLNQNVVHSILSKMMINQEIPAFWNESSKFILISKVNPTTLQNLALKLA 909
Query 122 ENVANAVEQNELTLNMKNPKFALTHERRFMRDGDRAGW 159
ENV +EQNEL LNMKNPKF ERR +++ W
Sbjct 910 ENVNEVMEQNELALNMKNPKFMFMQERRTQMKEEKSNW 947
> bbo:BBOV_III008380 17.m07733; eukaryotic translation initiation
factor 3 subunit 8; K03252 translation initiation factor
3 subunit C
Length=966
Score = 179 bits (455), Expect = 6e-45, Method: Compositional matrix adjust.
Identities = 88/189 (46%), Positives = 131/189 (69%), Gaps = 6/189 (3%)
Query 1 TVMAATKALQRGDWSDCARHIWSLSIWDKMFNKEHIQQMLLQKIKAEAMRTYIFTYLSLY 60
VM+ + LQ G+W +C + L+IW+++ ++E + Q+L +IK EA TYIF Y+ +Y
Sbjct 780 VVMSTFRHLQNGNWKECYDLLAGLTIWNRLPDREEVLQILKDRIKVEAFNTYIFKYVPVY 839
Query 61 DSFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASWDETSQFILISRVKHTRLQQLALTL 120
DSF++ QL+SMFDL N +HS++SKM+I +IHA+WD++S++ LI+ + T LQ+ A+ L
Sbjct 840 DSFSVDQLSSMFDLDNNVIHSLLSKMIITGDIHATWDDSSKYCLINHTEPTDLQKCAIKL 899
Query 121 AENVANAVEQNELTLNMKNPKFALTHERRFMRDGDRAGWGTRDREDGGGHRSRPGF-RGR 179
AEN+ AVEQNE+TLNMKNPKFAL+ +RRF ++ +G R R+D H + P F R R
Sbjct 900 AENLTTAVEQNEMTLNMKNPKFALSQDRRFQTRENKFSYG-RSRDD--RHHAAPTFNRNR 956
Query 180 AGLNPVGAR 188
+ GAR
Sbjct 957 RPIQ--GAR 963
> tpv:TP04_0739 eukaryotic translation initiation factor 3 subunit
8; K03252 translation initiation factor 3 subunit C
Length=951
Score = 178 bits (452), Expect = 1e-44, Method: Composition-based stats.
Identities = 85/166 (51%), Positives = 118/166 (71%), Gaps = 1/166 (0%)
Query 2 VMAATKALQRGDWSDCARHIWSLSIWDKMFNKEHIQQMLLQKIKAEAMRTYIFTYLSLYD 61
++ A K LQ GDW C I SL+ W+ M ++E +Q+ L + IK E RTYIF Y+++YD
Sbjct 759 ILNAFKHLQNGDWKQCFNFILSLNTWNNMPDREKVQETLKELIKVEGFRTYIFKYVNIYD 818
Query 62 SFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASWDETSQFILISRVKHTRLQQLALTLA 121
SF++ QL+SMF L N VHS++SKM++N EI SWD +S+ LI+ + T LQ+LA+ LA
Sbjct 819 SFSVEQLSSMFSLDENVVHSLISKMIVNGEILGSWDYSSKCCLINHSEPTELQKLAVKLA 878
Query 122 ENVANAVEQNELTLNMKNPKFALTHERRFMRDGDRAGWGTRDREDG 167
EN++ AVEQNELTLNMKN KFAL+ +RRF + R +G+R +DG
Sbjct 879 ENLSTAVEQNELTLNMKNSKFALSQDRRFQQRDTRYSYGSR-HDDG 923
> dre:334234 eif3c, eif3s8, fi37c04, wu:fb34a08, wu:fi37c04, zgc:66128;
eukaryotic translation initiation factor 3, subunit
C; K03252 translation initiation factor 3 subunit C
Length=926
Score = 129 bits (324), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 68/180 (37%), Positives = 110/180 (61%), Gaps = 20/180 (11%)
Query 2 VMAATKALQRGDWSDCARHI----WSLSIWDKMFNKEHIQQMLLQKIKAEAMRTYIFTYL 57
V+AA+KA++ GDW C I + +WD + +++ML++KI+ E++RTY+FTY
Sbjct 736 VVAASKAMKMGDWRTCHSFIINEKMNSKVWDLFPEAKCVREMLVRKIQEESLRTYLFTYS 795
Query 58 SLYDSFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASWDETSQFILISRVKHTRLQQLA 117
S+YDS ++ L+ MF+L L VHSI+SKM+INEE+ AS D+ +Q +++ R + T LQ +A
Sbjct 796 SVYDSISMETLSEMFELELPTVHSIISKMIINEELMASLDQPTQTVVMHRTEPTSLQNMA 855
Query 118 LTLAENVANAVEQNELTLNMKNPKFALTHERRFMRDGDRAGWGTRDREDGGGHRSRPGFR 177
L LAE + VE NE ++K G G+ RD++ GG++ + G++
Sbjct 856 LQLAEKLGGLVENNERVFDLKQ--------------GVYGGYFNRDQK--GGYQQKQGYQ 899
> hsa:8663 EIF3C, EIF3CL, EIF3S8, FLJ53378, FLJ54400, FLJ54404,
FLJ55450, FLJ55750, FLJ78287, MGC189737, MGC189744, eIF3-p110;
eukaryotic translation initiation factor 3, subunit C;
K03252 translation initiation factor 3 subunit C
Length=913
Score = 125 bits (313), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 71/184 (38%), Positives = 108/184 (58%), Gaps = 21/184 (11%)
Query 2 VMAATKALQRGDWSDCARHIWSLS----IWDKMFNKEHIQQMLLQKIKAEAMRTYIFTYL 57
V+AA+KA++ GDW C I + +WD + ++ ML++KI+ E++RTY+FTY
Sbjct 737 VVAASKAMKMGDWKTCHSFIINEKMNGKVWDLFPEADKVRTMLVRKIQEESLRTYLFTYS 796
Query 58 SLYDSFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASWDETSQFILISRVKHTRLQQLA 117
S+YDS ++ L+ MF+L L VHSI+SKM+INEE+ AS D+ +Q +++ R + T Q LA
Sbjct 797 SVYDSISMETLSDMFELDLPTVHSIISKMIINEELMASLDQPTQTVVMHRTEPTAQQNLA 856
Query 118 LTLAENVANAVEQNELTLNMKNPKFALTHERRFMRDGDRAGWGTRDREDGGGHRSRPGFR 177
L LAE + + VE NE + K G G+ RD++D G+R G+
Sbjct 857 LQLAEKLGSLVENNERVFDHKQ--------------GTYGGY-FRDQKD--GYRKNEGYM 899
Query 178 GRAG 181
R G
Sbjct 900 RRGG 903
> hsa:728689 EIF3CL; eukaryotic translation initiation factor
3, subunit C-like
Length=914
Score = 125 bits (313), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 71/184 (38%), Positives = 108/184 (58%), Gaps = 21/184 (11%)
Query 2 VMAATKALQRGDWSDCARHIWSLS----IWDKMFNKEHIQQMLLQKIKAEAMRTYIFTYL 57
V+AA+KA++ GDW C I + +WD + ++ ML++KI+ E++RTY+FTY
Sbjct 738 VVAASKAMKMGDWKTCHSFIINEKMNGKVWDLFPEADKVRTMLVRKIQEESLRTYLFTYS 797
Query 58 SLYDSFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASWDETSQFILISRVKHTRLQQLA 117
S+YDS ++ L+ MF+L L VHSI+SKM+INEE+ AS D+ +Q +++ R + T Q LA
Sbjct 798 SVYDSISMETLSDMFELDLPTVHSIISKMIINEELMASLDQPTQTVVMHRTEPTAQQNLA 857
Query 118 LTLAENVANAVEQNELTLNMKNPKFALTHERRFMRDGDRAGWGTRDREDGGGHRSRPGFR 177
L LAE + + VE NE + K G G+ RD++D G+R G+
Sbjct 858 LQLAEKLGSLVENNERVFDHKQ--------------GTYGGY-FRDQKD--GYRKNEGYM 900
Query 178 GRAG 181
R G
Sbjct 901 RRGG 904
> mmu:56347 Eif3c, 110kDa, 3230401O13Rik, Eif3s8, MGC36637, NIPIL(A3),
NipilA3; eukaryotic translation initiation factor 3,
subunit C; K03252 translation initiation factor 3 subunit
C
Length=911
Score = 125 bits (313), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 71/184 (38%), Positives = 108/184 (58%), Gaps = 21/184 (11%)
Query 2 VMAATKALQRGDWSDCARHIWSLS----IWDKMFNKEHIQQMLLQKIKAEAMRTYIFTYL 57
V+AA+KA++ GDW C I + +WD + ++ ML++KI+ E++RTY+FTY
Sbjct 735 VVAASKAMKMGDWKTCHSFIINEKMNGKVWDLFPEADKVRTMLVRKIQEESLRTYLFTYS 794
Query 58 SLYDSFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASWDETSQFILISRVKHTRLQQLA 117
S+YDS ++ L+ MF+L L VHSI+SKM+INEE+ AS D+ +Q +++ R + T Q LA
Sbjct 795 SVYDSISMETLSDMFELDLPTVHSIISKMIINEELMASLDQPTQTVVMHRTEPTAQQNLA 854
Query 118 LTLAENVANAVEQNELTLNMKNPKFALTHERRFMRDGDRAGWGTRDREDGGGHRSRPGFR 177
L LAE + + VE NE + K G G+ RD++D G+R G+
Sbjct 855 LQLAEKLGSLVENNERVFDHKQ--------------GTYGGY-FRDQKD--GYRKNEGYM 897
Query 178 GRAG 181
R G
Sbjct 898 RRGG 901
> ath:AT3G56150 EIF3C; EIF3C (EUKARYOTIC TRANSLATION INITIATION
FACTOR 3C); translation initiation factor; K03252 translation
initiation factor 3 subunit C
Length=900
Score = 124 bits (310), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 66/131 (50%), Positives = 91/131 (69%), Gaps = 0/131 (0%)
Query 2 VMAATKALQRGDWSDCARHIWSLSIWDKMFNKEHIQQMLLQKIKAEAMRTYIFTYLSLYD 61
VMAAT+AL +GD+ + SL +W + N++ I M+ +IK EA+RTY+FTY S Y+
Sbjct 668 VMAATRALTKGDFQKAFEVLNSLEVWRLLKNRDSILDMVKDRIKEEALRTYLFTYSSSYE 727
Query 62 SFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASWDETSQFILISRVKHTRLQQLALTLA 121
S ++ QLA MFD+S VHSIVSKMMINEE+HASWD+ ++ I+ V+H+RLQ LA L
Sbjct 728 SLSLDQLAKMFDVSEPQVHSIVSKMMINEELHASWDQPTRCIVFHEVQHSRLQSLAFQLT 787
Query 122 ENVANAVEQNE 132
E ++ E NE
Sbjct 788 EKLSILAESNE 798
> cpv:cgd4_3770 hypothetical protein ; K03252 translation initiation
factor 3 subunit C
Length=1040
Score = 122 bits (306), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 73/222 (32%), Positives = 119/222 (53%), Gaps = 25/222 (11%)
Query 1 TVMAATKALQRGDWSDCARHIWSLSIWDKMFNKEHIQQM---LLQKIKAEAMRTYIFTYL 57
T++AAT++LQ G W +C +++SLSIWD +N+E +QQ L IK EA+RTY+FTY
Sbjct 807 TIIAATRSLQLGKWRECRDYVFSLSIWD--YNQEDLQQTQERLELNIKQEALRTYLFTYG 864
Query 58 SLYDSFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASWDETSQFILISRVKHTRLQQLA 117
LY S+++ L MF L +HS++SKMM+ E+ A WD+T +F+L++ + +++Q +
Sbjct 865 HLYSSYSVNNLIEMFQLPREKIHSLLSKMMLKNELQALWDQTGEFVLLNHKQASKIQNDS 924
Query 118 LTLAENVANAVEQNELTLNM-------KNPKFALTHERRFMRDGDRAGWGTRDREDGGGH 170
L +A+ + V+ NE ++ KNP + + + AG G + + G G
Sbjct 925 LIVADKLLQFVDCNESMISSKGNASGGKNPN--------VINNRNSAGVGNPNNQGGIG- 975
Query 171 RSRPG-FRGRAGLNPVGARGRGLGRGTYRGCGFRGNRGSGDY 211
PG +G N G G ++ + SG Y
Sbjct 976 ---PGSVQGGGPNNTSNTNSSGTGNQSFNSNYYSRKTNSGQY 1014
> xla:100191027 eif3c, MGC114671, eif3-p110, eif3s8; eukaryotic
translation initiation factor 3, subunit C; K03252 translation
initiation factor 3 subunit C
Length=926
Score = 120 bits (302), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 61/141 (43%), Positives = 90/141 (63%), Gaps = 4/141 (2%)
Query 2 VMAATKALQRGDWSDCARHIWSLS----IWDKMFNKEHIQQMLLQKIKAEAMRTYIFTYL 57
V+AA+KA++ GDW C I + +WD E ++ ML++KI+ E++RTY+FTY
Sbjct 739 VVAASKAMKMGDWKTCKNFIINEKMNGKVWDLFPEAERVRGMLVRKIQEESLRTYLFTYS 798
Query 58 SLYDSFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASWDETSQFILISRVKHTRLQQLA 117
S+YDS +G L MF L + VHSI+SKM+INEE+ AS D+ +Q +++ + + LQ A
Sbjct 799 SVYDSIRMGILGDMFQLEIPTVHSIISKMIINEELMASLDQPTQTVVMHGTEPSSLQNTA 858
Query 118 LTLAENVANAVEQNELTLNMK 138
L LAE + N VE NE + K
Sbjct 859 LQLAEKLGNLVENNERIFDHK 879
> ath:AT3G22860 TIF3C2; TIF3C2; translation initiation factor;
K03252 translation initiation factor 3 subunit C
Length=805
Score = 116 bits (291), Expect = 6e-26, Method: Composition-based stats.
Identities = 63/151 (41%), Positives = 92/151 (60%), Gaps = 3/151 (1%)
Query 2 VMAATKALQRGDWSDCARHIWSLSIWDKMFNKEHIQQMLLQKIKAEAMRTYIFTYLS-LY 60
V+AAT+AL +G++ + + SL IW N++ I M+ I A+RTY+FTY S Y
Sbjct 626 VIAATRALIKGNFQEAFSVLNSLDIWRLFKNRDSILDMVKASISEVALRTYLFTYSSSCY 685
Query 61 DSFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASWDETSQFILISRVKHTRLQQLALTL 120
S ++ +LA MFD+S + V+SIVSKMMIN+E++A+WD+ +Q I+ V+H R Q LA +
Sbjct 686 KSLSLAELAKMFDISESHVYSIVSKMMINKELNATWDQPTQCIVFHEVQHNRAQSLAFQI 745
Query 121 AENVANAVEQNELTLNMKNPKFAL--THERR 149
E +A E NE + K L T RR
Sbjct 746 TEKLATLAESNESAMESKTGGVGLDMTSRRR 776
> cel:T23D8.4 eif-3.C; Eukaryotic Initiation Factor family member
(eif-3.C); K03252 translation initiation factor 3 subunit
C
Length=898
Score = 114 bits (285), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 53/138 (38%), Positives = 91/138 (65%), Gaps = 4/138 (2%)
Query 2 VMAATKALQRGDWSDCARHI----WSLSIWDKMFNKEHIQQMLLQKIKAEAMRTYIFTYL 57
V+AA+KA+ GDW C +I + +W+ N E ++ M++++I+ E++RTY+ TY
Sbjct 694 VVAASKAMLNGDWKKCQDYIVNDKMNQKVWNLFHNAETVKGMVVRRIQEESLRTYLLTYS 753
Query 58 SLYDSFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASWDETSQFILISRVKHTRLQQLA 117
++Y + ++ +LA +F+LS VHSI+SKM+I EE+ A+ DE + +++ RV+ +RLQ LA
Sbjct 754 TVYATVSLKKLADLFELSKKDVHSIISKMIIQEELSATLDEPTDCLIMHRVEPSRLQMLA 813
Query 118 LTLAENVANAVEQNELTL 135
L L++ + E NE L
Sbjct 814 LNLSDKLQTLAENNEQIL 831
> sce:YMR309C NIP1; Nip1p; K03252 translation initiation factor
3 subunit C
Length=812
Score = 73.9 bits (180), Expect = 5e-13, Method: Composition-based stats.
Identities = 36/130 (27%), Positives = 74/130 (56%), Gaps = 1/130 (0%)
Query 2 VMAATKALQRGDWSDCARHIWSLSIWDKMFNKEHIQQMLLQKIKAEAMRTYIFTYLSLYD 61
V+ A K++Q+G+W D +++ + W + N E + L ++++ E+++TY F++ Y
Sbjct 675 VLFAAKSMQKGNWRDSVKYLREIKSWALLPNMETVLNSLTERVQVESLKTYFFSFKRFYS 734
Query 62 SFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASW-DETSQFILISRVKHTRLQQLALTL 120
SF++ +LA +FDL N V ++ ++ EI A DE + F++ + T+L++ + L
Sbjct 735 SFSVAKLAELFDLPENKVVEVLQSVIAELEIPAKLNDEKTIFVVEKGDEITKLEEAMVKL 794
Query 121 AENVANAVEQ 130
+ A E+
Sbjct 795 NKEYKIAKER 804
> cel:K07H8.10 hypothetical protein; K11294 nucleolin
Length=798
Score = 38.5 bits (88), Expect = 0.025, Method: Composition-based stats.
Identities = 34/96 (35%), Positives = 45/96 (46%), Gaps = 15/96 (15%)
Query 147 ERRFMRDGDRAGWGTRDREDGGGHRSRPGFRGRA-GLNPVGARGRGLGRGTYRGCGFRGN 205
+R R GDR G R + G +R R F G + G N RG G G T RG
Sbjct 108 DRGNFRGGDRGGSPWRGGDRGVANRGRGDFSGGSRGGNKFSPRGGGRGGFTPRG------ 161
Query 206 RGSGDY-----RGDYCGESRGDLGEYRGYSDQRDPF 236
RG D+ RG++ +SRG G G+SD+R +
Sbjct 162 RGGNDFSPRGGRGNF--QSRGG-GSRVGFSDKRKQY 194
> hsa:1293 COL6A3, DKFZp686D23123, DKFZp686K04147, DKFZp686N0262,
FLJ34702, FLJ98399; collagen, type VI, alpha 3; K06238 collagen,
type VI, alpha
Length=3177
Score = 32.0 bits (71), Expect = 2.2, Method: Composition-based stats.
Identities = 25/71 (35%), Positives = 28/71 (39%), Gaps = 13/71 (18%)
Query 153 DGDRAGWGTRDREDGGGHRSRPGF------RGRAGLN----PVGARGRGLGRGTYRGCGF 202
D R G G+ R G R PG+ G GLN P G RGR RG G
Sbjct 2295 DDGRDGVGSEGRRGKKGERGFPGYPGPKGNPGEPGLNGTTGPKGIRGR---RGNSGPPGI 2351
Query 203 RGNRGSGDYRG 213
G +G Y G
Sbjct 2352 VGQKGDPGYPG 2362
> hsa:1278 COL1A2, OI4; collagen, type I, alpha 2; K06236 collagen,
type I/II/III/V/XI, alpha
Length=1366
Score = 31.6 bits (70), Expect = 3.0, Method: Composition-based stats.
Identities = 21/56 (37%), Positives = 25/56 (44%), Gaps = 2/56 (3%)
Query 152 RDGDRAGWGTRDREDGGGHRSRPGFRGRAGLNPVGARGRGLGRGTYRGCGFRGNRG 207
RDG+ G R+ GH+ G+ G G PVGA G G G GNRG
Sbjct 926 RDGNPGNDGPPGRDGQPGHKGERGYPGNIG--PVGAAGAPGPHGPVGPAGKHGNRG 979
Lambda K H
0.321 0.136 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8764825436
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40