bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0938_orf1
Length=172
Score E
Sequences producing significant alignments: (Bits) Value
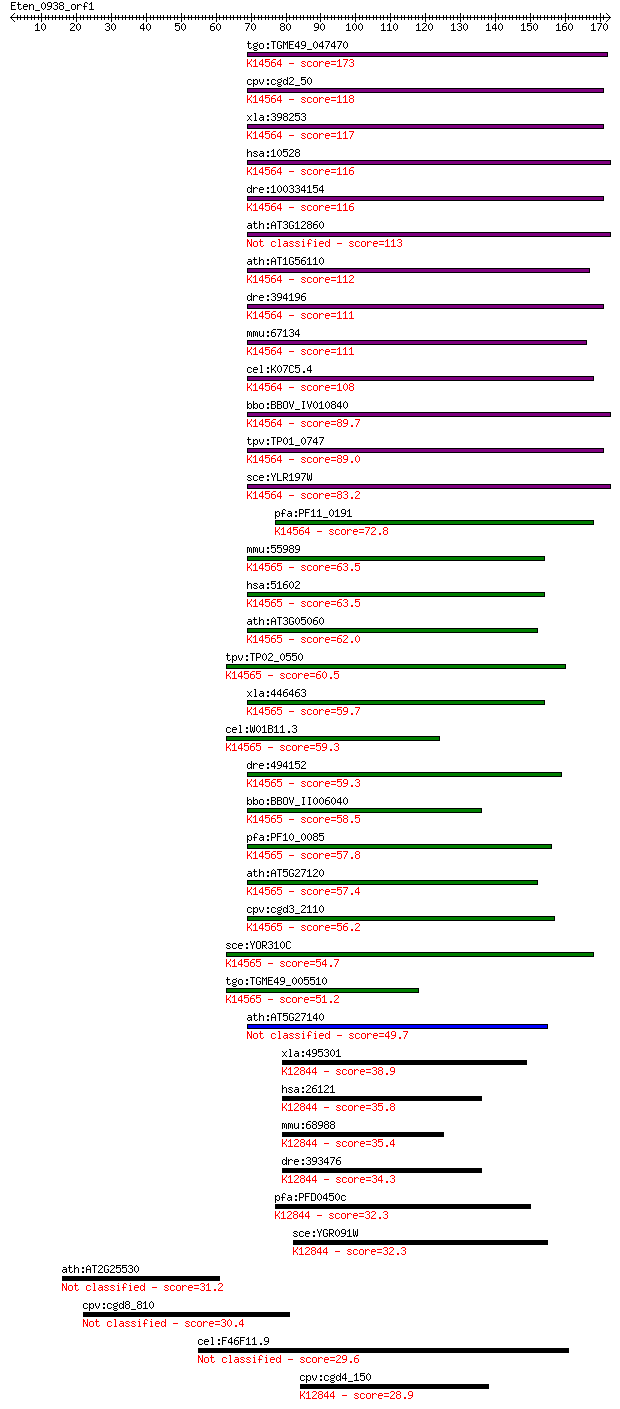
tgo:TGME49_047470 nucleolar protein 5A, putative ; K14564 nucl... 173 3e-43
cpv:cgd2_50 SIK1 nucleolar protein Nop56 ; K14564 nucleolar pr... 118 9e-27
xla:398253 nop56, MGC130628, nol5a, xnop56; NOP56 ribonucleopr... 117 1e-26
hsa:10528 NOP56, NOL5A; NOP56 ribonucleoprotein homolog (yeast... 116 4e-26
dre:100334154 NOP56-like; K14564 nucleolar protein 56 116 4e-26
ath:AT3G12860 nucleolar protein Nop56, putative 113 4e-25
ath:AT1G56110 NOP56; NOP56 (Arabidopsis homolog of nucleolar p... 112 8e-25
dre:394196 nop56, nol5a; NOP56 ribonucleoprotein homolog; K145... 111 1e-24
mmu:67134 Nop56, 2310044F10Rik, Nol5a; NOP56 ribonucleoprotein... 111 1e-24
cel:K07C5.4 hypothetical protein; K14564 nucleolar protein 56 108 1e-23
bbo:BBOV_IV010840 23.m05998; nucleolar protein Nop56; K14564 n... 89.7 5e-18
tpv:TP01_0747 hypothetical protein; K14564 nucleolar protein 56 89.0
sce:YLR197W NOP56, SIK1; Essential evolutionarily-conserved nu... 83.2 4e-16
pfa:PF11_0191 Nop56/Sik1-like protein, putative; K14564 nucleo... 72.8 5e-13
mmu:55989 Nop58, MGC105209, MSSP, Nol5, SIK, nop5; NOP58 ribon... 63.5 3e-10
hsa:51602 NOP58, NOP5, NOP5/NOP58; NOP58 ribonucleoprotein hom... 63.5 3e-10
ath:AT3G05060 SAR DNA-binding protein, putative; K14565 nucleo... 62.0 1e-09
tpv:TP02_0550 nuclear protein; K14565 nucleolar protein 58 60.5
xla:446463 nop58-a, MGC78950, nop5, nop5/nop58, nop58; NOP58 r... 59.7 4e-09
cel:W01B11.3 nol-5; NucleOLar protein family member (nol-5); K... 59.3 6e-09
dre:494152 nop58, NOP5/NOP58, nol5, wu:fa93b01, zgc:65841; NOP... 59.3 6e-09
bbo:BBOV_II006040 18.m06502; nucleolar protein NOP5; K14565 nu... 58.5 1e-08
pfa:PF10_0085 nucleolar protein NOP5, putative; K14565 nucleol... 57.8 2e-08
ath:AT5G27120 SAR DNA-binding protein, putative; K14565 nucleo... 57.4 2e-08
cpv:cgd3_2110 nucleolar protein NOP5/NOP58-like pre-mRNA splic... 56.2 6e-08
sce:YOR310C NOP58, NOP5; Nop58p; K14565 nucleolar protein 58 54.7
tgo:TGME49_005510 nucleolar protein NOP5, putative ; K14565 nu... 51.2 2e-06
ath:AT5G27140 SAR DNA-binding protein, putative 49.7 5e-06
xla:495301 prpf31, prp31, rp11; PRP31 pre-mRNA processing fact... 38.9 0.009
hsa:26121 PRPF31, DKFZp566J153, NY-BR-99, PRP31, RP11; PRP31 p... 35.8 0.067
mmu:68988 Prpf31, 1500019O16Rik, 2810404O06Rik, AW554706, PRP3... 35.4 0.093
dre:393476 prpf31, MGC66177, zgc:66177; PRP31 pre-mRNA process... 34.3 0.20
pfa:PFD0450c pre-mRNA splicing factor, putative; K12844 U4/U6 ... 32.3 0.72
sce:YGR091W PRP31; Prp31p; K12844 U4/U6 small nuclear ribonucl... 32.3 0.76
ath:AT2G25530 AFG1-like ATPase family protein 31.2 2.0
cpv:cgd8_810 hypothetical protein 30.4 3.3
cel:F46F11.9 hypothetical protein 29.6 5.3
cpv:cgd4_150 pre-mRNA splicing protein; Prp31p--like ; K12844 ... 28.9 7.9
> tgo:TGME49_047470 nucleolar protein 5A, putative ; K14564 nucleolar
protein 56
Length=536
Score = 173 bits (438), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 82/103 (79%), Positives = 94/103 (91%), Gaps = 0/103 (0%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK+++GRTPKYGLLFHSSFIGRVQ+Q+HRGRMSRYLASKCALAARIDAF+DE +
Sbjct 355 RALKSKNGRTPKYGLLFHSSFIGRVQKQQHRGRMSRYLASKCALAARIDAFADEETPESA 414
Query 129 DQPRQHVFGDKLREQLEERLKYLSDGIVPRKNLDVMREAAAEV 171
D R +V+G KLREQLEERLKYL+DGIVPRKNLDVMREAA+E+
Sbjct 415 DGIRSNVYGVKLREQLEERLKYLADGIVPRKNLDVMREAASEL 457
> cpv:cgd2_50 SIK1 nucleolar protein Nop56 ; K14564 nucleolar
protein 56
Length=499
Score = 118 bits (296), Expect = 9e-27, Method: Compositional matrix adjust.
Identities = 57/102 (55%), Positives = 79/102 (77%), Gaps = 11/102 (10%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK++ G TPKYGLLF S++IG+ Q K++GR+SRYLA+KC++AARID FS
Sbjct 343 RALKSK-GNTPKYGLLFQSTYIGKASQ-KNKGRISRYLANKCSIAARIDNFS-------- 392
Query 129 DQPRQHVFGDKLREQLEERLKYLSDGIVPRKNLDVMREAAAE 170
++FG+KL++Q+E+RLKYLS+GI P KN+D+MREA +E
Sbjct 393 -TVNNNIFGEKLKQQVEDRLKYLSEGISPPKNIDIMREAISE 433
> xla:398253 nop56, MGC130628, nol5a, xnop56; NOP56 ribonucleoprotein
homolog; K14564 nucleolar protein 56
Length=532
Score = 117 bits (294), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 62/102 (60%), Positives = 74/102 (72%), Gaps = 11/102 (10%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK R G TPKYGL+FHS+FIGR K++GR+SRYLA+KC +A+RID FS+ P
Sbjct 337 RALKTR-GNTPKYGLIFHSTFIGRAAM-KNKGRISRYLANKCTIASRIDCFSE---IPTS 391
Query 129 DQPRQHVFGDKLREQLEERLKYLSDGIVPRKNLDVMREAAAE 170
VFGDKLREQ+EERL + G VPRKNLDVM+EA E
Sbjct 392 ------VFGDKLREQVEERLAFYETGEVPRKNLDVMKEAQQE 427
> hsa:10528 NOP56, NOL5A; NOP56 ribonucleoprotein homolog (yeast);
K14564 nucleolar protein 56
Length=594
Score = 116 bits (291), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 60/104 (57%), Positives = 76/104 (73%), Gaps = 11/104 (10%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK R G TPKYGL+FHS+FIGR K++GR+SRYLA+KC++A+RID FS+ P
Sbjct 337 RALKTR-GNTPKYGLIFHSTFIGRAAA-KNKGRISRYLANKCSIASRIDCFSE---VPTS 391
Query 129 DQPRQHVFGDKLREQLEERLKYLSDGIVPRKNLDVMREAAAEVE 172
VFG+KLREQ+EERL + G +PRKNLDVM+EA + E
Sbjct 392 ------VFGEKLREQVEERLSFYETGEIPRKNLDVMKEAMVQAE 429
> dre:100334154 NOP56-like; K14564 nucleolar protein 56
Length=432
Score = 116 bits (290), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 60/102 (58%), Positives = 74/102 (72%), Gaps = 11/102 (10%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK R G TPKYGL+FHS+FIGR K++GR+SRYLA+KC +A+RID FS+ P
Sbjct 222 RALKTR-GNTPKYGLIFHSTFIGRAAA-KNKGRISRYLANKCTIASRIDCFSE---VPTS 276
Query 129 DQPRQHVFGDKLREQLEERLKYLSDGIVPRKNLDVMREAAAE 170
VFGDKLR+Q+EERL + G PRKNLDVM+EA A+
Sbjct 277 ------VFGDKLRDQVEERLAFYETGEAPRKNLDVMKEAVAQ 312
> ath:AT3G12860 nucleolar protein Nop56, putative
Length=499
Score = 113 bits (282), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 56/104 (53%), Positives = 74/104 (71%), Gaps = 11/104 (10%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK R G TPKYGL+FHSSFIGR K++GR++R+LA+KC++A+RID FSD +
Sbjct 340 RALKTR-GNTPKYGLIFHSSFIGRASA-KNKGRIARFLANKCSIASRIDCFSDNSTT--- 394
Query 129 DQPRQHVFGDKLREQLEERLKYLSDGIVPRKNLDVMREAAAEVE 172
FG+KLREQ+EERL + G+ PRKN+DVM+E +E
Sbjct 395 ------AFGEKLREQVEERLDFYDKGVAPRKNVDVMKEVLENLE 432
> ath:AT1G56110 NOP56; NOP56 (Arabidopsis homolog of nucleolar
protein Nop56); K14564 nucleolar protein 56
Length=522
Score = 112 bits (279), Expect = 8e-25, Method: Composition-based stats.
Identities = 56/98 (57%), Positives = 73/98 (74%), Gaps = 11/98 (11%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK R G TPKYGL+FHSSFIGR K++GR++RYLA+KC++A+RID F+D A
Sbjct 340 RALKTR-GNTPKYGLIFHSSFIGRASA-KNKGRIARYLANKCSIASRIDCFADGATT--- 394
Query 129 DQPRQHVFGDKLREQLEERLKYLSDGIVPRKNLDVMRE 166
FG+KLREQ+EERL++ G+ PRKN+DVM+E
Sbjct 395 ------AFGEKLREQVEERLEFYDKGVAPRKNVDVMKE 426
> dre:394196 nop56, nol5a; NOP56 ribonucleoprotein homolog; K14564
nucleolar protein 56
Length=494
Score = 111 bits (278), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 59/102 (57%), Positives = 72/102 (70%), Gaps = 11/102 (10%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK R G TPKYGL+FHS+FIGR K++GR+SRYLA+KC +A+RID FS+ P
Sbjct 338 RALKTR-GNTPKYGLIFHSTFIGRAAA-KNKGRISRYLANKCTIASRIDCFSE---VPTS 392
Query 129 DQPRQHVFGDKLREQLEERLKYLSDGIVPRKNLDVMREAAAE 170
VFGDKL Q+EERL + G PRKNLDVM+EA A+
Sbjct 393 ------VFGDKLVTQVEERLAFYETGEAPRKNLDVMKEAVAQ 428
> mmu:67134 Nop56, 2310044F10Rik, Nol5a; NOP56 ribonucleoprotein
homolog (yeast); K14564 nucleolar protein 56
Length=580
Score = 111 bits (278), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 57/97 (58%), Positives = 72/97 (74%), Gaps = 11/97 (11%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK R G TPKYGL+FHS+FIGR K++GR+SRYLA+KC++A+RID FS+ P
Sbjct 337 RALKTR-GNTPKYGLIFHSTFIGRAAA-KNKGRISRYLANKCSIASRIDCFSE---VPTS 391
Query 129 DQPRQHVFGDKLREQLEERLKYLSDGIVPRKNLDVMR 165
VFG+KLREQ+EERL + G +PRKNLDVM+
Sbjct 392 ------VFGEKLREQVEERLSFYETGEIPRKNLDVMK 422
> cel:K07C5.4 hypothetical protein; K14564 nucleolar protein 56
Length=486
Score = 108 bits (270), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 55/99 (55%), Positives = 74/99 (74%), Gaps = 11/99 (11%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK RS TPKYGLLFHSSFIG+ K++GR+SRYLA+KC++AAR+D FS+ P
Sbjct 343 RALKTRSN-TPKYGLLFHSSFIGKAGT-KNKGRVSRYLANKCSIAARVDCFSE---TPVS 397
Query 129 DQPRQHVFGDKLREQLEERLKYLSDGIVPRKNLDVMREA 167
+G+ LR+Q+E+RL+Y + G VP+KN+DVM+EA
Sbjct 398 ------TYGEFLRQQVEDRLEYFTSGTVPKKNIDVMKEA 430
> bbo:BBOV_IV010840 23.m05998; nucleolar protein Nop56; K14564
nucleolar protein 56
Length=569
Score = 89.7 bits (221), Expect = 5e-18, Method: Composition-based stats.
Identities = 48/104 (46%), Positives = 68/104 (65%), Gaps = 11/104 (10%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK RS TPKYGL+F S+FIG+ KH+GR +RYLA+KCALAAR+D F D
Sbjct 338 RALKTRSN-TPKYGLIFQSTFIGKASV-KHKGRAARYLANKCALAARLDCFCD------- 388
Query 129 DQPRQHVFGDKLREQLEERLKYLSDGIVPRKNLDVMREAAAEVE 172
+V+G + + L +R++YL+ G V +++VM+ A+ E E
Sbjct 389 --VNSNVYGKHMVDLLAKRMEYLAGGPVHETSIEVMKAASKEYE 430
> tpv:TP01_0747 hypothetical protein; K14564 nucleolar protein
56
Length=560
Score = 89.0 bits (219), Expect = 7e-18, Method: Composition-based stats.
Identities = 49/102 (48%), Positives = 66/102 (64%), Gaps = 11/102 (10%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK+R+ TPKYGLLF S+FIG+ K +G+ +RYLA+KCALAAR+D F D
Sbjct 336 RALKSRTN-TPKYGLLFQSTFIGKASN-KLKGKAARYLANKCALAARLDYFCD------- 386
Query 129 DQPRQHVFGDKLREQLEERLKYLSDGIVPRKNLDVMREAAAE 170
V+G K+ EQL +R+ YL G KN++VM++A E
Sbjct 387 --VNTDVYGKKMSEQLTKRMDYLLGGPQHDKNINVMKQAHEE 426
> sce:YLR197W NOP56, SIK1; Essential evolutionarily-conserved
nucleolar protein component of the box C/D snoRNP complexes
that direct 2'-O-methylation of pre-rRNA during its maturation;
overexpression causes spindle orientation defects; K14564
nucleolar protein 56
Length=504
Score = 83.2 bits (204), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 50/113 (44%), Positives = 74/113 (65%), Gaps = 20/113 (17%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK + G TPKYGL++HS FI + K++GR+SRYLA+KC++A+RID +S+E P+
Sbjct 344 RALKTK-GNTPKYGLIYHSGFISKASA-KNKGRISRYLANKCSMASRIDNYSEE---PS- 397
Query 129 DQPRQHVFGDKLREQLEERLKYLSDGIVPRKNLDVMREA---------AAEVE 172
+VFG L++Q+E+RL++ + G KN ++EA AAEVE
Sbjct 398 -----NVFGSVLKKQVEQRLEFYNTGKPTLKNELAIQEAMELYNKDKPAAEVE 445
> pfa:PF11_0191 Nop56/Sik1-like protein, putative; K14564 nucleolar
protein 56
Length=594
Score = 72.8 bits (177), Expect = 5e-13, Method: Composition-based stats.
Identities = 37/91 (40%), Positives = 59/91 (64%), Gaps = 10/91 (10%)
Query 77 RTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAGDQPRQHVF 136
+TPKYG+L++SS+I + Q +GRMSRYL+ K A+AARID+FSD + +
Sbjct 346 KTPKYGILYNSSYISKTPIQL-KGRMSRYLSCKSAMAARIDSFSDYP---------TNSY 395
Query 137 GDKLREQLEERLKYLSDGIVPRKNLDVMREA 167
G ++QLE +++++ G+ KN+D + EA
Sbjct 396 GLIFKKQLEHKIQHMVKGVKLSKNIDYINEA 426
> mmu:55989 Nop58, MGC105209, MSSP, Nol5, SIK, nop5; NOP58 ribonucleoprotein
homolog (yeast); K14565 nucleolar protein 58
Length=536
Score = 63.5 bits (153), Expect = 3e-10, Method: Composition-based stats.
Identities = 36/85 (42%), Positives = 52/85 (61%), Gaps = 11/85 (12%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK+R TPKYGL++H+S +G+ KH+G++SR LA+K LA R DAF +++ G
Sbjct 327 RALKSRRD-TPKYGLIYHASLVGQ-SSPKHKGKISRMLAAKTVLAIRYDAFGEDSSSAMG 384
Query 129 DQPRQHVFGDKLREQLEERLKYLSD 153
+ R +LE RL+ L D
Sbjct 385 IENRA---------KLEARLRILED 400
> hsa:51602 NOP58, NOP5, NOP5/NOP58; NOP58 ribonucleoprotein homolog
(yeast); K14565 nucleolar protein 58
Length=529
Score = 63.5 bits (153), Expect = 3e-10, Method: Composition-based stats.
Identities = 36/85 (42%), Positives = 52/85 (61%), Gaps = 11/85 (12%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK+R TPKYGL++H+S +G+ KH+G++SR LA+K LA R DAF +++ G
Sbjct 327 RALKSRRD-TPKYGLIYHASLVGQTSP-KHKGKISRMLAAKTVLAIRYDAFGEDSSSAMG 384
Query 129 DQPRQHVFGDKLREQLEERLKYLSD 153
+ R +LE RL+ L D
Sbjct 385 VENRA---------KLEARLRTLED 400
> ath:AT3G05060 SAR DNA-binding protein, putative; K14565 nucleolar
protein 58
Length=533
Score = 62.0 bits (149), Expect = 1e-09, Method: Composition-based stats.
Identities = 36/83 (43%), Positives = 49/83 (59%), Gaps = 11/83 (13%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK + TPKYGL+FH+S +G+ KH+G++SR LA+K LA R+DA D D G
Sbjct 326 RALKTKHA-TPKYGLIFHASLVGQAAP-KHKGKISRSLAAKTVLAIRVDALGDSQDNTMG 383
Query 129 DQPRQHVFGDKLREQLEERLKYL 151
+ R +LE RL+ L
Sbjct 384 LENRA---------KLEARLRNL 397
> tpv:TP02_0550 nuclear protein; K14565 nucleolar protein 58
Length=425
Score = 60.5 bits (145), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 39/98 (39%), Positives = 57/98 (58%), Gaps = 11/98 (11%)
Query 63 GGEGGV-RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSD 121
G E + RALK RS TPKYG+++H+ +G+ KH+G++SR LA+K AL R+DA
Sbjct 323 GAEKALFRALKTRS-HTPKYGIIYHAGLVGQ-SSPKHKGKISRILAAKLALCVRVDALG- 379
Query 122 EADQPAGDQPRQHVFGDKLREQLEERLKYLSDGIVPRK 159
E+D+P + +KL ++ LSDG RK
Sbjct 380 ESDKPTVALENKKYVENKL-------VQLLSDGNQKRK 410
> xla:446463 nop58-a, MGC78950, nop5, nop5/nop58, nop58; NOP58
ribonucleoprotein homolog; K14565 nucleolar protein 58
Length=534
Score = 59.7 bits (143), Expect = 4e-09, Method: Composition-based stats.
Identities = 34/85 (40%), Positives = 52/85 (61%), Gaps = 11/85 (12%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK R TPKYGL++H+S +G+ K++G++SR LA+K ALA R DA ++ +
Sbjct 328 RALKTRKD-TPKYGLIYHASLVGQT-TPKNKGKISRMLAAKAALAIRYDALGEDTNAE-- 383
Query 129 DQPRQHVFGDKLREQLEERLKYLSD 153
G + R +LE RL++L +
Sbjct 384 -------LGVETRAKLESRLRHLEE 401
> cel:W01B11.3 nol-5; NucleOLar protein family member (nol-5);
K14565 nucleolar protein 58
Length=487
Score = 59.3 bits (142), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 31/62 (50%), Positives = 43/62 (69%), Gaps = 3/62 (4%)
Query 63 GGEGGV-RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSD 121
G E + RALK + TPKYGL++H+ I + K +G+M+R LA+KC+LA RIDA SD
Sbjct 318 GAEKALFRALKTKKD-TPKYGLIYHAQLITQAPP-KVKGKMARKLAAKCSLATRIDALSD 375
Query 122 EA 123
E+
Sbjct 376 ES 377
> dre:494152 nop58, NOP5/NOP58, nol5, wu:fa93b01, zgc:65841; NOP58
ribonucleoprotein homolog (yeast); K14565 nucleolar protein
58
Length=529
Score = 59.3 bits (142), Expect = 6e-09, Method: Composition-based stats.
Identities = 35/90 (38%), Positives = 54/90 (60%), Gaps = 11/90 (12%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK R TPKYGL++H+S +G+ K++G++SR LA+K ALA R DA ++ + G
Sbjct 328 RALKTRRD-TPKYGLIYHASLVGQT-TAKNKGKISRMLAAKTALAIRYDALGEDTNAEMG 385
Query 129 DQPRQHVFGDKLREQLEERLKYLSDGIVPR 158
+ R +LE RL++L + + R
Sbjct 386 VENRA---------KLEARLRFLEEKGIRR 406
> bbo:BBOV_II006040 18.m06502; nucleolar protein NOP5; K14565
nucleolar protein 58
Length=439
Score = 58.5 bits (140), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 45/67 (67%), Gaps = 2/67 (2%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK + TPKYG+++H+ F+G+ Q KH+G++SR LA+K AL R+DA D +
Sbjct 330 RALKT-NAPTPKYGIIYHAGFVGQAQP-KHKGKISRILAAKLALCVRVDALQDSQEPTVA 387
Query 129 DQPRQHV 135
+ ++++
Sbjct 388 IESKRYL 394
> pfa:PF10_0085 nucleolar protein NOP5, putative; K14565 nucleolar
protein 58
Length=469
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/87 (35%), Positives = 52/87 (59%), Gaps = 11/87 (12%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK +S +TPKYGL++H++ +G+ K +GR+SR LA+K +L R+DA + +
Sbjct 330 RALKTKS-KTPKYGLIYHATLVGQT-APKLKGRISRSLAAKLSLCTRVDALGNFVEPSIA 387
Query 129 DQPRQHVFGDKLREQLEERLKYLSDGI 155
+ H LE+RL+Y++ +
Sbjct 388 ITCKSH---------LEKRLEYITSNL 405
> ath:AT5G27120 SAR DNA-binding protein, putative; K14565 nucleolar
protein 58
Length=533
Score = 57.4 bits (137), Expect = 2e-08, Method: Composition-based stats.
Identities = 35/83 (42%), Positives = 48/83 (57%), Gaps = 11/83 (13%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK + TPKYGL+FH+S +G+ K++G++SR LA+K LA R DA D D G
Sbjct 325 RALKTKHA-TPKYGLIFHASVVGQA-APKNKGKISRSLAAKSVLAIRCDALGDSQDNTMG 382
Query 129 DQPRQHVFGDKLREQLEERLKYL 151
+ R +LE RL+ L
Sbjct 383 VENRL---------KLEARLRTL 396
> cpv:cgd3_2110 nucleolar protein NOP5/NOP58-like pre-mRNA splicinig
factor prp31 ; K14565 nucleolar protein 58
Length=467
Score = 56.2 bits (134), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 31/88 (35%), Positives = 53/88 (60%), Gaps = 11/88 (12%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
RALK + TPKYGL++H++ +G+ K +G++SR LA+K +L R+DA +D+ +
Sbjct 333 RALKTKKS-TPKYGLIYHAAVVGQ-SAPKLKGKISRILAAKLSLCIRVDALNDQNEPTVA 390
Query 129 DQPRQHVFGDKLREQLEERLKYLSDGIV 156
+ +Q+V E RL+ LS+ +
Sbjct 391 IENKQYV---------ERRLEELSNQLT 409
> sce:YOR310C NOP58, NOP5; Nop58p; K14565 nucleolar protein 58
Length=511
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 41/106 (38%), Positives = 64/106 (60%), Gaps = 11/106 (10%)
Query 63 GGEGGV-RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSD 121
G E + RALK + TPKYGLL+H+S +G+ K++G+++R LA+K A++ R DA ++
Sbjct 321 GAEKALFRALKTKHD-TPKYGLLYHASLVGQA-TGKNKGKIARVLAAKAAVSLRYDALAE 378
Query 122 EADQPAGDQPRQHVFGDKLREQLEERLKYLSDGIVPRKNLDVMREA 167
+ D +GD G + R ++E RL L +G R V+REA
Sbjct 379 DRDD-SGD------IGLESRAKVENRLSQL-EGRDLRTTPKVVREA 416
> tgo:TGME49_005510 nucleolar protein NOP5, putative ; K14565
nucleolar protein 58
Length=490
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 27/56 (48%), Positives = 39/56 (69%), Gaps = 3/56 (5%)
Query 63 GGEGGV-RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARID 117
G E + RALK +S TPKYGLLFH++ +G+ K +G++SR LA+K +L R+D
Sbjct 324 GAEKALFRALKTKS-HTPKYGLLFHAALVGQA-PPKLKGKISRVLAAKLSLCVRVD 377
> ath:AT5G27140 SAR DNA-binding protein, putative
Length=445
Score = 49.7 bits (117), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 32/86 (37%), Positives = 49/86 (56%), Gaps = 11/86 (12%)
Query 69 RALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAG 128
+ALK + TPKYGL++H+ + R +++G+++R LA+K ALA R DAF + D G
Sbjct 295 KALKTKQA-TPKYGLIYHAPLV-RQAAPENKGKIARSLAAKSALAIRCDAFGNGQDNTMG 352
Query 129 DQPRQHVFGDKLREQLEERLKYLSDG 154
+ R +LE RL+ L G
Sbjct 353 VESRL---------KLEARLRNLEGG 369
> xla:495301 prpf31, prp31, rp11; PRP31 pre-mRNA processing factor
31 homolog; K12844 U4/U6 small nuclear ribonucleoprotein
PRP31
Length=498
Score = 38.9 bits (89), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 37/70 (52%), Gaps = 10/70 (14%)
Query 79 PKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAGDQPRQHVFGD 138
P G ++HS + + HR + +R +++KC LA+R+D+F + P G G
Sbjct 268 PHTGYIYHSEIVQSLPSDLHR-KAARLVSAKCTLASRVDSFH---ENPEGK------IGY 317
Query 139 KLREQLEERL 148
L+E++E +
Sbjct 318 DLKEEIERKF 327
> hsa:26121 PRPF31, DKFZp566J153, NY-BR-99, PRP31, RP11; PRP31
pre-mRNA processing factor 31 homolog (S. cerevisiae); K12844
U4/U6 small nuclear ribonucleoprotein PRP31
Length=499
Score = 35.8 bits (81), Expect = 0.067, Method: Compositional matrix adjust.
Identities = 16/57 (28%), Positives = 31/57 (54%), Gaps = 1/57 (1%)
Query 79 PKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAGDQPRQHV 135
P G ++HS + + R + +R +A+KC LAAR+D+F + + G + + +
Sbjct 269 PHTGYIYHSDIVQSLPPDLRR-KAARLVAAKCTLAARVDSFHESTEGKVGYELKDEI 324
> mmu:68988 Prpf31, 1500019O16Rik, 2810404O06Rik, AW554706, PRP31,
RP11; PRP31 pre-mRNA processing factor 31 homolog (yeast);
K12844 U4/U6 small nuclear ribonucleoprotein PRP31
Length=493
Score = 35.4 bits (80), Expect = 0.093, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 27/46 (58%), Gaps = 1/46 (2%)
Query 79 PKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEAD 124
P G ++HS + + R + +R +A+KC LAAR+D+F + +
Sbjct 269 PHTGYIYHSDIVQSLPPDLRR-KAARLVAAKCTLAARVDSFHESTE 313
> dre:393476 prpf31, MGC66177, zgc:66177; PRP31 pre-mRNA processing
factor 31 homolog (yeast); K12844 U4/U6 small nuclear
ribonucleoprotein PRP31
Length=508
Score = 34.3 bits (77), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 15/57 (26%), Positives = 31/57 (54%), Gaps = 1/57 (1%)
Query 79 PKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAGDQPRQHV 135
P G ++H + + R + +R +++KC LA+R+D+F + AD G ++ +
Sbjct 280 PHTGYIYHCDVVQTLPPDLRR-KAARLVSAKCTLASRVDSFHESADGKVGYDLKEEI 335
> pfa:PFD0450c pre-mRNA splicing factor, putative; K12844 U4/U6
small nuclear ribonucleoprotein PRP31
Length=534
Score = 32.3 bits (72), Expect = 0.72, Method: Composition-based stats.
Identities = 24/73 (32%), Positives = 35/73 (47%), Gaps = 1/73 (1%)
Query 77 RTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAGDQPRQHVF 136
+T G+L S + V + +S LASKC+LAARID F + G RQ++
Sbjct 327 KTLGIGILCCSEIVQSVPDAYKKKAIS-LLASKCSLAARIDYFKKYKEGQYGLLLRQYII 385
Query 137 GDKLREQLEERLK 149
++ Q LK
Sbjct 386 SHLIKLQEPPPLK 398
> sce:YGR091W PRP31; Prp31p; K12844 U4/U6 small nuclear ribonucleoprotein
PRP31
Length=494
Score = 32.3 bits (72), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 37/73 (50%), Gaps = 8/73 (10%)
Query 82 GLLFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAGDQPRQHVFGDKLR 141
G LF S I + H+ +M R L +K +LAAR+D A Q GD R V K +
Sbjct 283 GYLFASDMIQKFPVSVHK-QMLRMLCAKVSLAARVD-----AGQKNGD--RNTVLAHKWK 334
Query 142 EQLEERLKYLSDG 154
+L ++ + LS+
Sbjct 335 AELSKKARKLSEA 347
> ath:AT2G25530 AFG1-like ATPase family protein
Length=655
Score = 31.2 bits (69), Expect = 2.0, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 24/45 (53%), Gaps = 5/45 (11%)
Query 16 VPEQQNGCGVAQFEGSCGRSVGGSAYIACRGFSVARQVPSLYNSD 60
VPE +G FE CGR VG + YIA VA+ +++ SD
Sbjct 488 VPESCSGVARFTFEYLCGRPVGAADYIA-----VAKNYHTIFISD 527
> cpv:cgd8_810 hypothetical protein
Length=1342
Score = 30.4 bits (67), Expect = 3.3, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 26/59 (44%), Gaps = 4/59 (6%)
Query 22 GCGVAQFEGSCGRSVGGSAYIACRGFSVARQVPSLYNSDSRGGEGGVRALKARSGRTPK 80
G G A GSCGRS + + C V + + D+RG GG R R R+ +
Sbjct 657 GNGAAGNSGSCGRSTCVKSSVEC----VCKYNSNGIYEDNRGSGGGARCRTGRKRRSSR 711
> cel:F46F11.9 hypothetical protein
Length=1280
Score = 29.6 bits (65), Expect = 5.3, Method: Composition-based stats.
Identities = 31/116 (26%), Positives = 47/116 (40%), Gaps = 15/116 (12%)
Query 55 SLYNSDSR---GGEGGVRALKARSGRTPKYGLLFHSSFIGRVQQQKHRGRMSRYLASKCA 111
+LYN+ S G A +TP++ +L H S R + R LA CA
Sbjct 143 TLYNTTSTLMIPGHCSTPKWAAPHAKTPRHYILLHDSRSPRSSTE----RRDELLAQMCA 198
Query 112 LAA-------RIDAFSDEADQPAGDQPRQHVFGDKLREQLEERLKYLSDGIVPRKN 160
++D+ S+ A+ G F D L + LEE ++ +D I P N
Sbjct 199 TYGNDNCQMLQLDSDSESAEM-KGVWDEIDEFNDVLEKGLEEAHQHSTDAIAPGPN 253
> cpv:cgd4_150 pre-mRNA splicing protein; Prp31p--like ; K12844
U4/U6 small nuclear ribonucleoprotein PRP31
Length=463
Score = 28.9 bits (63), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 25/54 (46%), Gaps = 0/54 (0%)
Query 84 LFHSSFIGRVQQQKHRGRMSRYLASKCALAARIDAFSDEADQPAGDQPRQHVFG 137
+ S I R Q K++ + R ++ KC L ARID S + G R ++
Sbjct 285 IISQSDIVRNIQDKYKKKAIRLVSLKCGLCARIDFSSTDKSPDHGVNYRNYILN 338
Lambda K H
0.318 0.133 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4341553636
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40