bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0935_orf1
Length=173
Score E
Sequences producing significant alignments: (Bits) Value
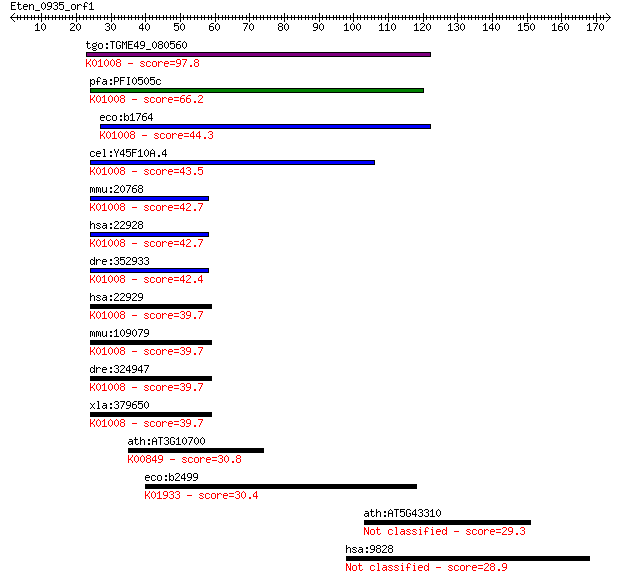
tgo:TGME49_080560 selenophosphate synthetase, putative (EC:2.7... 97.8 2e-20
pfa:PFI0505c selenide water dikinase, putative (EC:2.7.9.3); K... 66.2 5e-11
eco:b1764 selD, ECK1762, fdhB, JW1753; selenophosphate synthas... 44.3 2e-04
cel:Y45F10A.4 seld-1; SELD (SelD homolog) involved in selenoph... 43.5 3e-04
mmu:20768 Sephs2, Sps2, Ysg3; selenophosphate synthetase 2 (EC... 42.7 6e-04
hsa:22928 SEPHS2, SPS2; selenophosphate synthetase 2 (EC:2.7.9... 42.7 7e-04
dre:352933 sps2, MGC92096, cb701, zgc:92096; selenophosphate s... 42.4 8e-04
hsa:22929 SEPHS1, MGC4980, SELD, SPS, SPS1; selenophosphate sy... 39.7 0.005
mmu:109079 Sephs1, 1110046B24Rik, AA589574, AI505014, AW111620... 39.7 0.006
dre:324947 sephs1, wu:fc49b09, zgc:55304; selenophosphate synt... 39.7 0.006
xla:379650 sephs1, MGC68922; selenophosphate synthetase 1 (EC:... 39.7 0.006
ath:AT3G10700 GHMP kinase family protein (EC:2.7.1.6); K00849 ... 30.8 2.3
eco:b2499 purM, ECK2495, JW2484, purG, purI; phosphoribosylami... 30.4 3.5
ath:AT5G43310 COP1-interacting protein-related 29.3 6.6
hsa:9828 ARHGEF17, FLJ90019, KIAA0337, P164RHOGEF, TEM4, p164-... 28.9 8.0
> tgo:TGME49_080560 selenophosphate synthetase, putative (EC:2.7.9.3
1.8.1.4); K01008 selenide, water dikinase [EC:2.7.9.3]
Length=1189
Score = 97.8 bits (242), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 50/99 (50%), Positives = 65/99 (65%), Gaps = 1/99 (1%)
Query 23 LLAQSVDFFRLFTGDLYTMGRVAAAHALSDLYACGAEPLTALAVTLMQKAPPELQANNLL 82
LL Q+VDF+R F D + +GR+AA HA+SD YA GAEPL AL ++ + + ANNLL
Sbjct 779 LLIQTVDFYRAFFDDAFVLGRIAATHAMSDCYAMGAEPLVALLTAVVPYSTDCIMANNLL 838
Query 83 QFLCGASSVFSSEGCELSGGHSAAGDGPSAAGFCVTGRI 121
Q L G S S + C+L+GGHSA G AAG +TGR+
Sbjct 839 QLLGGCCSALSRDRCQLAGGHSAQGR-DMAAGLTITGRL 876
> pfa:PFI0505c selenide water dikinase, putative (EC:2.7.9.3);
K01008 selenide, water dikinase [EC:2.7.9.3]
Length=1193
Score = 66.2 bits (160), Expect = 5e-11, Method: Composition-based stats.
Identities = 35/96 (36%), Positives = 50/96 (52%), Gaps = 1/96 (1%)
Query 24 LAQSVDFFRLFTGDLYTMGRVAAAHALSDLYACGAEPLTALAVTLMQKAPPELQANNLLQ 83
L Q++DFF+ F D Y +G + A H+LSD+Y+ G + AL V +++ L
Sbjct 758 LVQTIDFFKSFIDDEYILGSIIAIHSLSDIYSMGGTGICALCVLIVKDNIERKLQQRLEN 817
Query 84 FLCGASSVFSSEGCELSGGHSAAGDGPSAAGFCVTG 119
L G E C LSGGH+ AG+ + G VTG
Sbjct 818 VLTGCCQKLKEERCVLSGGHTCAGN-ENYVGLAVTG 852
> eco:b1764 selD, ECK1762, fdhB, JW1753; selenophosphate synthase
(EC:2.7.9.3); K01008 selenide, water dikinase [EC:2.7.9.3]
Length=347
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 31/98 (31%), Positives = 48/98 (48%), Gaps = 9/98 (9%)
Query 27 SVDFFRLFTGDLYTMGRVAAAHALSDLYACGAEPLTALAV--TLMQKAPPELQANNLLQF 84
+ DFF + + GR+AA +A+SD++A G +P+ A+A+ + K PE+ +
Sbjct 66 TTDFFMPIVDNPFDFGRIAATNAISDIFAMGGKPIMAIAILGWPINKLSPEIAR----EV 121
Query 85 LCGASSVFSSEGCELSGGHSAAGDGPSAA-GFCVTGRI 121
G G L+GGHS D P G VTG +
Sbjct 122 TEGGRYACRQAGIALAGGHSI--DAPEPIFGLAVTGIV 157
> cel:Y45F10A.4 seld-1; SELD (SelD homolog) involved in selenophosphate
synthesis family member (seld-1); K01008 selenide,
water dikinase [EC:2.7.9.3]
Length=378
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 30/89 (33%), Positives = 42/89 (47%), Gaps = 12/89 (13%)
Query 24 LAQSVDFFRLFTGDLYTMGRVAAAHALSDLYACGAEP------LTALAVTLMQKAPPELQ 77
L Q+ DFF D Y MGRV A+ LSDLYA G L A+A+ L +K Q
Sbjct 78 LVQTTDFFYPLIDDPYIMGRVTCANVLSDLYAMGVSECDNMLMLLAVAIDLNEK-----Q 132
Query 78 ANNLL-QFLCGASSVFSSEGCELSGGHSA 105
+ ++ F+ G G ++ GG +
Sbjct 133 RDIVVPLFIQGFKDAADEAGTKIRGGQTV 161
> mmu:20768 Sephs2, Sps2, Ysg3; selenophosphate synthetase 2 (EC:2.7.9.3);
K01008 selenide, water dikinase [EC:2.7.9.3]
Length=452
Score = 42.7 bits (99), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 19/34 (55%), Positives = 22/34 (64%), Gaps = 0/34 (0%)
Query 24 LAQSVDFFRLFTGDLYTMGRVAAAHALSDLYACG 57
L Q+ DFF D Y MGR+A A+ LSDLYA G
Sbjct 136 LVQTTDFFYPLVEDPYMMGRIACANVLSDLYAMG 169
> hsa:22928 SEPHS2, SPS2; selenophosphate synthetase 2 (EC:2.7.9.3);
K01008 selenide, water dikinase [EC:2.7.9.3]
Length=448
Score = 42.7 bits (99), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 19/34 (55%), Positives = 22/34 (64%), Gaps = 0/34 (0%)
Query 24 LAQSVDFFRLFTGDLYTMGRVAAAHALSDLYACG 57
L Q+ DFF D Y MGR+A A+ LSDLYA G
Sbjct 133 LVQTTDFFYPLVEDPYMMGRIACANVLSDLYAMG 166
> dre:352933 sps2, MGC92096, cb701, zgc:92096; selenophosphate
synthetase 2 (EC:2.7.9.3); K01008 selenide, water dikinase
[EC:2.7.9.3]
Length=447
Score = 42.4 bits (98), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 19/34 (55%), Positives = 22/34 (64%), Gaps = 0/34 (0%)
Query 24 LAQSVDFFRLFTGDLYTMGRVAAAHALSDLYACG 57
L Q+ DFF D Y MGR+A A+ LSDLYA G
Sbjct 118 LVQTTDFFYPLVEDPYMMGRIACANVLSDLYAMG 151
> hsa:22929 SEPHS1, MGC4980, SELD, SPS, SPS1; selenophosphate
synthetase 1 (EC:2.7.9.3); K01008 selenide, water dikinase [EC:2.7.9.3]
Length=325
Score = 39.7 bits (91), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 17/35 (48%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 24 LAQSVDFFRLFTGDLYTMGRVAAAHALSDLYACGA 58
L Q+ D+ D Y MGR+A A+ LSDLYA G
Sbjct 15 LVQTTDYIYPIVDDPYMMGRIACANVLSDLYAMGV 49
> mmu:109079 Sephs1, 1110046B24Rik, AA589574, AI505014, AW111620,
SELD, SPS, SPS1; selenophosphate synthetase 1 (EC:2.7.9.3);
K01008 selenide, water dikinase [EC:2.7.9.3]
Length=392
Score = 39.7 bits (91), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 17/35 (48%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 24 LAQSVDFFRLFTGDLYTMGRVAAAHALSDLYACGA 58
L Q+ D+ D Y MGR+A A+ LSDLYA G
Sbjct 82 LVQTTDYIYPIVDDPYMMGRIACANVLSDLYAMGV 116
> dre:324947 sephs1, wu:fc49b09, zgc:55304; selenophosphate synthetase
1 (EC:2.7.9.3); K01008 selenide, water dikinase [EC:2.7.9.3]
Length=392
Score = 39.7 bits (91), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 17/35 (48%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 24 LAQSVDFFRLFTGDLYTMGRVAAAHALSDLYACGA 58
L Q+ D+ D Y MGR+A A+ LSDLYA G
Sbjct 82 LVQTTDYIYPIVDDPYMMGRIACANVLSDLYAMGV 116
> xla:379650 sephs1, MGC68922; selenophosphate synthetase 1 (EC:2.7.9.3);
K01008 selenide, water dikinase [EC:2.7.9.3]
Length=392
Score = 39.7 bits (91), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 17/35 (48%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 24 LAQSVDFFRLFTGDLYTMGRVAAAHALSDLYACGA 58
L Q+ D+ D Y MGR+A A+ LSDLYA G
Sbjct 82 LVQTTDYIYPIVDDPYMMGRIACANVLSDLYAMGV 116
> ath:AT3G10700 GHMP kinase family protein (EC:2.7.1.6); K00849
galactokinase [EC:2.7.1.6]
Length=424
Score = 30.8 bits (68), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 18/41 (43%), Positives = 25/41 (60%), Gaps = 3/41 (7%)
Query 35 TGDLYTMGRVAAAHALSDL--YACGAEPLTALAVTLMQKAP 73
+G+L G++ +A LS + Y CGAEPL L L+ KAP
Sbjct 319 SGNLEEFGKLISASGLSSIENYECGAEPLIQLYKILL-KAP 358
> eco:b2499 purM, ECK2495, JW2484, purG, purI; phosphoribosylaminoimidazole
synthetase (EC:6.3.3.1); K01933 phosphoribosylformylglycinamidine
cyclo-ligase [EC:6.3.3.1]
Length=345
Score = 30.4 bits (67), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 26/84 (30%), Positives = 35/84 (41%), Gaps = 10/84 (11%)
Query 40 TMGRVAAAHALSDLYACGAEPLTALAVTLMQKAPPELQANNLLQFLCGASSVFSSEGCEL 99
T+G A ++DL GAEPL L A +L + + G + GC L
Sbjct 82 TIGIDLVAMCVNDLVVQGAEPLFFLD----YYATGKLDVDTASAVISGIAEGCLQSGCSL 137
Query 100 SGGHSAA------GDGPSAAGFCV 117
GG +A G+ AGFCV
Sbjct 138 VGGETAEMPGMYHGEDYDVAGFCV 161
> ath:AT5G43310 COP1-interacting protein-related
Length=1237
Score = 29.3 bits (64), Expect = 6.6, Method: Composition-based stats.
Identities = 15/53 (28%), Positives = 26/53 (49%), Gaps = 5/53 (9%)
Query 103 HSAAGDGPSAAGFCVTGRIFYPSPPGASS-----AKTNSSKGGNDEKRRQQQQ 150
HS G P G+ + G +YP PGAS T+ S+ G+ +++ ++
Sbjct 368 HSPPGTFPVFQGYTMQGMPYYPGYPGASPYPSPYPSTDDSRRGSGQRKARKHH 420
> hsa:9828 ARHGEF17, FLJ90019, KIAA0337, P164RHOGEF, TEM4, p164-RhoGEF;
Rho guanine nucleotide exchange factor (GEF) 17
Length=2063
Score = 28.9 bits (63), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 34/77 (44%), Gaps = 9/77 (11%)
Query 98 ELSGGHSAAGDG-------PSAAGFCVTGRIFYPSPPGASSAKTNSSKGGNDEKRRQQQQ 150
ELSG S+ D P A FC G P +SSA+TN G ++
Sbjct 635 ELSGPESSLTDEGIGADPEPPVAAFCGLGTTGMWRPLSSSSAQTNHHGPGTEDS--LGGW 692
Query 151 ALVAPGKPLAPGFLLRK 167
ALV+P P PG L R+
Sbjct 693 ALVSPETPPTPGALRRR 709
Lambda K H
0.316 0.133 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4406352944
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40