bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0926_orf1
Length=92
Score E
Sequences producing significant alignments: (Bits) Value
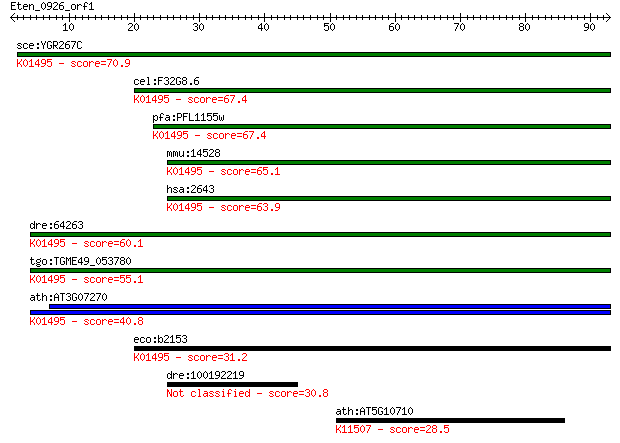
sce:YGR267C FOL2; Fol2p (EC:3.5.4.16); K01495 GTP cyclohydrola... 70.9 9e-13
cel:F32G8.6 cat-4; abnormal CATecholamine distribution family ... 67.4 1e-11
pfa:PFL1155w GTP-CH; GTP cyclohydrolase I (EC:3.5.4.16); K0149... 67.4 1e-11
mmu:14528 Gch1, GTP-CH, GTPCH, Gch; GTP cyclohydrolase 1 (EC:3... 65.1 6e-11
hsa:2643 GCH1, DYT14, DYT5, DYT5a, GCH, GTP-CH-1, GTPCH1, HPAB... 63.9 1e-10
dre:64263 gch2, fb51g02, gch, gch1b, wu:fb51g02; GTP cyclohydr... 60.1 2e-09
tgo:TGME49_053780 GTP cyclohydrolase I, putative (EC:3.5.4.16)... 55.1 6e-08
ath:AT3G07270 GTP cyclohydrolase I (EC:3.5.4.16); K01495 GTP c... 40.8 0.001
eco:b2153 folE, ECK2146, JW2140; GTP cyclohydrolase I (EC:3.5.... 31.2 0.86
dre:100192219 gch1, zgc:195269; GTP cyclohydrolase 1 (EC:3.5.4... 30.8 0.94
ath:AT5G10710 protein binding; K11507 centromere protein O 28.5 5.1
> sce:YGR267C FOL2; Fol2p (EC:3.5.4.16); K01495 GTP cyclohydrolase
I [EC:3.5.4.16]
Length=243
Score = 70.9 bits (172), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 35/92 (38%), Positives = 60/92 (65%), Gaps = 4/92 (4%)
Query 2 KRMEQAVSDLLKALPAGVLSEEVLLNTPRRAAEAFLYFTKGYELS-LEAAVGSGVFAYTE 60
+R+ A+ +L L V + E LL+TP+R A+A LYFTKGY+ + ++ + + VF E
Sbjct 59 QRISGAIKTILTELGEDV-NREGLLDTPQRYAKAMLYFTKGYQTNIMDDVIKNAVFE--E 115
Query 61 ETGDTIVIKDLFVHSVCEHHLLPFHGTCSIAY 92
+ + ++++D+ ++S+CEHHL+PF G I Y
Sbjct 116 DHDEMVIVRDIEIYSLCEHHLVPFFGKVHIGY 147
> cel:F32G8.6 cat-4; abnormal CATecholamine distribution family
member (cat-4); K01495 GTP cyclohydrolase I [EC:3.5.4.16]
Length=223
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 47/73 (64%), Gaps = 2/73 (2%)
Query 20 LSEEVLLNTPRRAAEAFLYFTKGYELSLEAAVGSGVFAYTEETGDTIVIKDLFVHSVCEH 79
++ + LL TP RAA+A + FTKGY+ L+ + VF E+ + +++KD+ + S+CEH
Sbjct 58 INRQGLLKTPERAAKAMMAFTKGYDDQLDELLNEAVF--DEDHDEMVIVKDIEMFSLCEH 115
Query 80 HLLPFHGTCSIAY 92
HL+PF G I Y
Sbjct 116 HLVPFMGKVHIGY 128
> pfa:PFL1155w GTP-CH; GTP cyclohydrolase I (EC:3.5.4.16); K01495
GTP cyclohydrolase I [EC:3.5.4.16]
Length=389
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 28/70 (40%), Positives = 41/70 (58%), Gaps = 0/70 (0%)
Query 23 EVLLNTPRRAAEAFLYFTKGYELSLEAAVGSGVFAYTEETGDTIVIKDLFVHSVCEHHLL 82
++L T RR AE FLY T GY L +E + ++ + I + + ++S+C+HHLL
Sbjct 223 DILKRTNRRYAETFLYLTNGYNLDIEQIIKRSLYKRMYKNNSIIKVTGIHIYSLCKHHLL 282
Query 83 PFHGTCSIAY 92
PF GTC I Y
Sbjct 283 PFEGTCDIEY 292
> mmu:14528 Gch1, GTP-CH, GTPCH, Gch; GTP cyclohydrolase 1 (EC:3.5.4.16);
K01495 GTP cyclohydrolase I [EC:3.5.4.16]
Length=241
Score = 65.1 bits (157), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 29/68 (42%), Positives = 43/68 (63%), Gaps = 2/68 (2%)
Query 25 LLNTPRRAAEAFLYFTKGYELSLEAAVGSGVFAYTEETGDTIVIKDLFVHSVCEHHLLPF 84
LL TP RAA A YFTKGY+ ++ + +F E+ + +++KD+ + S+CEHHL+PF
Sbjct 82 LLKTPWRAATAMQYFTKGYQETISDVLNDAIF--DEDHDEMVIVKDIDMFSMCEHHLVPF 139
Query 85 HGTCSIAY 92
G I Y
Sbjct 140 VGRVHIGY 147
> hsa:2643 GCH1, DYT14, DYT5, DYT5a, GCH, GTP-CH-1, GTPCH1, HPABH4B;
GTP cyclohydrolase 1 (EC:3.5.4.16); K01495 GTP cyclohydrolase
I [EC:3.5.4.16]
Length=250
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 28/68 (41%), Positives = 43/68 (63%), Gaps = 2/68 (2%)
Query 25 LLNTPRRAAEAFLYFTKGYELSLEAAVGSGVFAYTEETGDTIVIKDLFVHSVCEHHLLPF 84
LL TP RAA A +FTKGY+ ++ + +F E+ + +++KD+ + S+CEHHL+PF
Sbjct 91 LLKTPWRAASAMQFFTKGYQETISDVLNDAIF--DEDHDEMVIVKDIDMFSMCEHHLVPF 148
Query 85 HGTCSIAY 92
G I Y
Sbjct 149 VGKVHIGY 156
> dre:64263 gch2, fb51g02, gch, gch1b, wu:fb51g02; GTP cyclohydrolase
2 (EC:3.5.4.16); K01495 GTP cyclohydrolase I [EC:3.5.4.16]
Length=240
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 31/89 (34%), Positives = 51/89 (57%), Gaps = 3/89 (3%)
Query 4 MEQAVSDLLKALPAGVLSEEVLLNTPRRAAEAFLYFTKGYELSLEAAVGSGVFAYTEETG 63
+E A + +L+ L + LL TP RAA+A + TKGY ++ + +F E+
Sbjct 59 LEAAYTTILRGLGENT-DRQGLLKTPLRAAKAMQFLTKGYHETIYDILNDAIF--DEDHE 115
Query 64 DTIVIKDLFVHSVCEHHLLPFHGTCSIAY 92
+ +++KD+ + S+CEHHL+PF G I Y
Sbjct 116 ELVIVKDIDMFSLCEHHLVPFFGKVHIGY 144
> tgo:TGME49_053780 GTP cyclohydrolase I, putative (EC:3.5.4.16);
K01495 GTP cyclohydrolase I [EC:3.5.4.16]
Length=563
Score = 55.1 bits (131), Expect = 6e-08, Method: Composition-based stats.
Identities = 30/98 (30%), Positives = 50/98 (51%), Gaps = 10/98 (10%)
Query 4 MEQAVSDLLKALPAGVLSEEVLLNTPRRAAEAFLYFTKGYELSLEAAVGSGVFAYTEETG 63
+E+ V +L+A+ V + L TP+R A A +F++GY + + + +F+ E
Sbjct 367 IEEGVRMILRAMGEDV-TRAGLRETPKRVAAAMEFFSRGYRADPKDVIRNALFSVEEGHE 425
Query 64 D---------TIVIKDLFVHSVCEHHLLPFHGTCSIAY 92
D + + + + S+CEHHLLPF G C I Y
Sbjct 426 DGQYLAGKQGMVTVGRIDISSLCEHHLLPFFGKCHIGY 463
> ath:AT3G07270 GTP cyclohydrolase I (EC:3.5.4.16); K01495 GTP
cyclohydrolase I [EC:3.5.4.16]
Length=466
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 32/98 (32%), Positives = 48/98 (48%), Gaps = 12/98 (12%)
Query 7 AVSDLLKALPAGV---LSEEVLLNTPRRAAEAFLYFTKGYELSLEAAVGSGVFA------ 57
A+ D +K L G+ ++ E + TP R A+A T+GY+ ++ V S +F
Sbjct 34 AIQDAVKLLLQGLHEDVNREGIKKTPFRVAKALREGTRGYKQKVKDYVQSALFPEAGLDE 93
Query 58 ---YTEETGDTIVIKDLFVHSVCEHHLLPFHGTCSIAY 92
G +V++DL +S CE LLPFH C I Y
Sbjct 94 GVGQAGGVGGLVVVRDLDHYSYCESCLLPFHVKCHIGY 131
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 31/90 (34%), Positives = 44/90 (48%), Gaps = 3/90 (3%)
Query 4 MEQAVSDLLKALPAGVLSEEVLLNTPRRAAEAFLYFTK-GYELSLEAAVGSGVFAYTEET 62
M AV +LK+L L +E L+ TP R + L F + E+ L + + V +E
Sbjct 270 MVSAVVSILKSLGEDPLRKE-LIATPTRFLKWMLNFQRTNLEMKLNSFNPAKVNGEVKEK 328
Query 63 GDTIVIKDLFVHSVCEHHLLPFHGTCSIAY 92
+ F S+CEHHLLPF+G I Y
Sbjct 329 RLHCELNMPF-WSMCEHHLLPFYGVVHIGY 357
> eco:b2153 folE, ECK2146, JW2140; GTP cyclohydrolase I (EC:3.5.4.16);
K01495 GTP cyclohydrolase I [EC:3.5.4.16]
Length=222
Score = 31.2 bits (69), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 17/74 (22%), Positives = 36/74 (48%), Gaps = 3/74 (4%)
Query 20 LSEEVLLNTPRRAAEAFLYFTKGYE-LSLEAAVGSGVFAYTEETGDTIVIKDLFVHSVCE 78
L+++ L+ TP R A+ +Y + + L + + + + ++D+ + S CE
Sbjct 55 LADDSLMETPHRIAK--MYVDEIFSGLDYANFPKITLIENKMKVDEMVTVRDITLTSTCE 112
Query 79 HHLLPFHGTCSIAY 92
HH + G ++AY
Sbjct 113 HHFVTIDGKATVAY 126
> dre:100192219 gch1, zgc:195269; GTP cyclohydrolase 1 (EC:3.5.4.16)
Length=104
Score = 30.8 bits (68), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 13/20 (65%), Positives = 15/20 (75%), Gaps = 0/20 (0%)
Query 25 LLNTPRRAAEAFLYFTKGYE 44
LL TP RAA A +FTKGY+
Sbjct 63 LLKTPWRAATAMQFFTKGYQ 82
> ath:AT5G10710 protein binding; K11507 centromere protein O
Length=312
Score = 28.5 bits (62), Expect = 5.1, Method: Composition-based stats.
Identities = 14/36 (38%), Positives = 21/36 (58%), Gaps = 1/36 (2%)
Query 51 VGSGVFAYTEETGDTIVIKDLFVHSVCE-HHLLPFH 85
VG + AY + +IK+LF H + E +H LP+H
Sbjct 184 VGDLLQAYVDRKEQVRLIKELFGHQISEIYHSLPYH 219
Lambda K H
0.321 0.135 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2003222032
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40