bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0922_orf4
Length=166
Score E
Sequences producing significant alignments: (Bits) Value
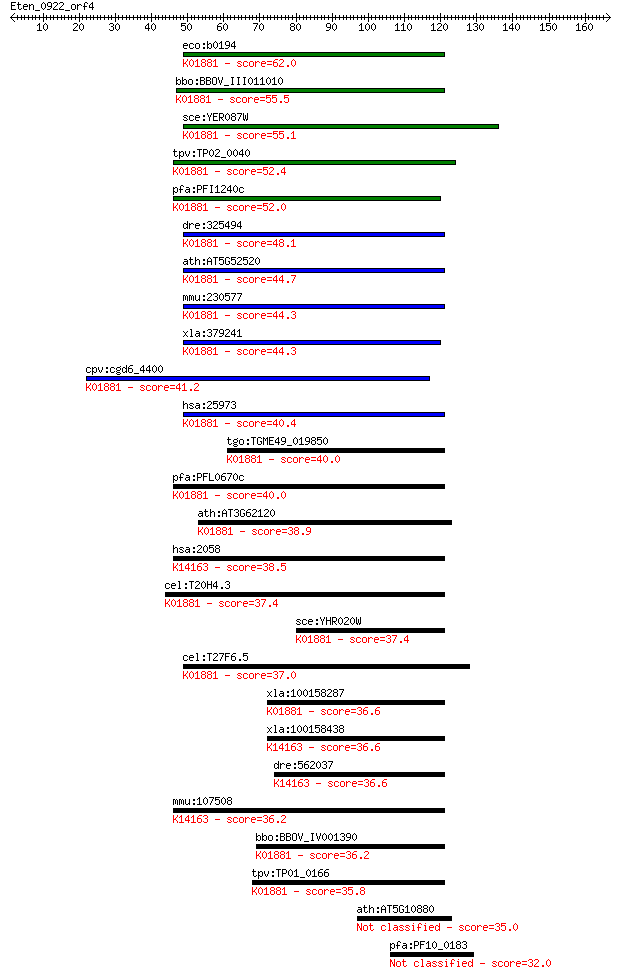
eco:b0194 proS, drpA, ECK0194, JW0190; prolyl-tRNA synthetase ... 62.0 8e-10
bbo:BBOV_III011010 17.m07948; tRNA synthetase class II core do... 55.5 8e-08
sce:YER087W AIM10; Aim10p (EC:6.1.1.15); K01881 prolyl-tRNA sy... 55.1 1e-07
tpv:TP02_0040 prolyl-tRNA synthetase; K01881 prolyl-tRNA synth... 52.4 7e-07
pfa:PFI1240c prolyl-t-RNA synthase, putative (EC:6.1.1.15); K0... 52.0 1e-06
dre:325494 pars2, fc83e06, wu:fc83e06, zgc:77163; prolyl-tRNA ... 48.1 1e-05
ath:AT5G52520 OVA6; OVA6 (OVULE ABORTION 6); ATP binding / ami... 44.7 1e-04
mmu:230577 Pars2, BC027073, MGC47008; prolyl-tRNA synthetase (... 44.3 2e-04
xla:379241 pars2, MGC53152; prolyl-tRNA synthetase 2, mitochon... 44.3 2e-04
cpv:cgd6_4400 proline-tRNA synthetase; class II aaRS (ybak RNA... 41.2 0.001
hsa:25973 PARS2, DKFZp727A071, MGC14416, MGC19467, MT-PRORS; p... 40.4 0.003
tgo:TGME49_019850 prolyl-tRNA synthetase, putative (EC:6.1.1.1... 40.0 0.003
pfa:PFL0670c bifunctional aminoacyl-tRNA synthetase, putative ... 40.0 0.004
ath:AT3G62120 tRNA synthetase class II (G, H, P and S) family ... 38.9 0.009
hsa:2058 EPRS, DKFZp313B047, EARS, GLUPRORS, PARS, QARS, QPRS;... 38.5 0.012
cel:T20H4.3 prs-1; Prolyl tRNA Synthetase family member (prs-1... 37.4 0.025
sce:YHR020W Protein of unknown function that may interact with... 37.4 0.026
cel:T27F6.5 prs-2; Prolyl tRNA Synthetase family member (prs-2... 37.0 0.027
xla:100158287 hypothetical protein LOC100158287; K01881 prolyl... 36.6 0.036
xla:100158438 eprs; glutamyl-prolyl-tRNA synthetase; K14163 bi... 36.6 0.040
dre:562037 eprs, wu:fb38d08, wu:ft34d10; glutamyl-prolyl-tRNA ... 36.6 0.043
mmu:107508 Eprs, 2410081F06Rik, 3010002K18Rik, C79379, Qprs; g... 36.2 0.046
bbo:BBOV_IV001390 21.m02889; prolyl-tRNA synthetase (EC:6.1.1.... 36.2 0.055
tpv:TP01_0166 prolyl-tRNA synthetase; K01881 prolyl-tRNA synth... 35.8 0.075
ath:AT5G10880 tRNA synthetase-related / tRNA ligase-related 35.0 0.12
pfa:PF10_0183 eukaryotic translation initiation factor subunit... 32.0 0.93
> eco:b0194 proS, drpA, ECK0194, JW0190; prolyl-tRNA synthetase
(EC:6.1.1.15); K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=572
Score = 62.0 bits (149), Expect = 8e-10, Method: Composition-based stats.
Identities = 28/72 (38%), Positives = 44/72 (61%), Gaps = 0/72 (0%)
Query 49 LEVGHCFQLPKAYCKAARARFMDSQGVEKVPFMSSYGLGVSRLIAHLALSHQDMKGLLFP 108
+EVGH FQL Y +A +A G ++ M YG+GV+R++A + D +G+++P
Sbjct 409 IEVGHIFQLGTKYSEALKASVQGEDGRNQILTMGCYGIGVTRVVAAAIEQNYDERGIVWP 468
Query 109 PQVAPFCVAIIP 120
+APF VAI+P
Sbjct 469 DAIAPFQVAILP 480
> bbo:BBOV_III011010 17.m07948; tRNA synthetase class II core
domain containing protein; K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=439
Score = 55.5 bits (132), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 28/75 (37%), Positives = 45/75 (60%), Gaps = 2/75 (2%)
Query 47 TCLEVGHCFQLPKAYCKAARARFMDSQGVEKVP-FMSSYGLGVSRLIAHLALSHQDMKGL 105
T +E+ H FQL A A ++ + +G K P +++SYG+G+ RL+ A H D +G+
Sbjct 235 TLMEIAHIFQLGDAITTEAGLKY-EGEGRMKHPVYLNSYGIGIHRLLQAAASQHADSEGI 293
Query 106 LFPPQVAPFCVAIIP 120
P +AP+ VA+IP
Sbjct 294 RLPQIIAPYDVAVIP 308
> sce:YER087W AIM10; Aim10p (EC:6.1.1.15); K01881 prolyl-tRNA
synthetase [EC:6.1.1.15]
Length=576
Score = 55.1 bits (131), Expect = 1e-07, Method: Composition-based stats.
Identities = 32/88 (36%), Positives = 46/88 (52%), Gaps = 5/88 (5%)
Query 49 LEVGHCFQLPKAYCKAARARFMDSQGV-EKVPFMSSYGLGVSRLIAHLALSHQDMKGLLF 107
+EVGH F L Y K +F+D + E M YG+GVSRL+ +A +D G +
Sbjct 379 IEVGHIFLLGNKYSKPLNVKFVDKENKNETFVHMGCYGIGVSRLVGAIAELGRDSNGFRW 438
Query 108 PPQVAPFCVAIIPQPAARWNDGRNAARL 135
P +AP+ V+I P N+ N+ RL
Sbjct 439 PAIMAPYKVSICTGP----NNPENSQRL 462
> tpv:TP02_0040 prolyl-tRNA synthetase; K01881 prolyl-tRNA synthetase
[EC:6.1.1.15]
Length=461
Score = 52.4 bits (124), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 40/78 (51%), Gaps = 0/78 (0%)
Query 46 GTCLEVGHCFQLPKAYCKAARARFMDSQGVEKVPFMSSYGLGVSRLIAHLALSHQDMKGL 105
T LEVGH F+ Y +F S + +M+SYG+G++RL+ L ++ D G+
Sbjct 261 NTELEVGHLFKFGTHYSDVYGLKFEISGRKREKLYMNSYGIGINRLLNVLINNYSDEFGI 320
Query 106 LFPPQVAPFCVAIIPQPA 123
P +AP+ V II
Sbjct 321 KLPQLIAPYDVTIIDSTT 338
> pfa:PFI1240c prolyl-t-RNA synthase, putative (EC:6.1.1.15);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=579
Score = 52.0 bits (123), Expect = 1e-06, Method: Composition-based stats.
Identities = 26/74 (35%), Positives = 39/74 (52%), Gaps = 0/74 (0%)
Query 46 GTCLEVGHCFQLPKAYCKAARARFMDSQGVEKVPFMSSYGLGVSRLIAHLALSHQDMKGL 105
G E H F+L Y +++D + +K M SYG+G+ RL+ L + D +G+
Sbjct 295 GKYKEAAHIFKLGDYYSNKLDIKYLDKKNEKKNILMGSYGIGIYRLLYFLIENFYDEEGI 354
Query 106 LFPPQVAPFCVAII 119
P QVAPF V +I
Sbjct 355 KLPQQVAPFSVYLI 368
> dre:325494 pars2, fc83e06, wu:fc83e06, zgc:77163; prolyl-tRNA
synthetase (mitochondrial) (EC:6.1.1.15); K01881 prolyl-tRNA
synthetase [EC:6.1.1.15]
Length=477
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 24/72 (33%), Positives = 40/72 (55%), Gaps = 0/72 (0%)
Query 49 LEVGHCFQLPKAYCKAARARFMDSQGVEKVPFMSSYGLGVSRLIAHLALSHQDMKGLLFP 108
+EVGH F L Y +A +A F+++ V V M +GLGV+R++A + +P
Sbjct 307 IEVGHTFYLGTKYSQAFKALFVNASNVPAVAEMGCFGLGVTRILAASIEVMSTEDAIHWP 366
Query 109 PQVAPFCVAIIP 120
+AP+ V ++P
Sbjct 367 GLIAPYQVCVLP 378
> ath:AT5G52520 OVA6; OVA6 (OVULE ABORTION 6); ATP binding / aminoacyl-tRNA
ligase/ nucleotide binding / proline-tRNA ligase;
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=543
Score = 44.7 bits (104), Expect = 1e-04, Method: Composition-based stats.
Identities = 20/72 (27%), Positives = 39/72 (54%), Gaps = 1/72 (1%)
Query 49 LEVGHCFQLPKAYCKAARARFMDSQGVEKVPFMSSYGLGVSRLIAHLALSHQDMKGLLFP 108
L+ G L + + +A +F D G + + +S+ + +R + + ++H D GL+ P
Sbjct 283 LQAGTSHNLGQNFSRAFGTQFADENGERQHVWQTSWAVS-TRFVGGIIMTHGDDTGLMLP 341
Query 109 PQVAPFCVAIIP 120
P++AP V I+P
Sbjct 342 PKIAPIQVVIVP 353
> mmu:230577 Pars2, BC027073, MGC47008; prolyl-tRNA synthetase
(mitochondrial)(putative) (EC:6.1.1.15); K01881 prolyl-tRNA
synthetase [EC:6.1.1.15]
Length=511
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 40/74 (54%), Gaps = 4/74 (5%)
Query 49 LEVGHCFQLPKAYCKAARARFMDSQGVEKVPFMSSYGLGVSRLIAHL--ALSHQDMKGLL 106
+EVGH F L Y A F ++ G + M YGLGV+R++A LS +D +
Sbjct 341 IEVGHTFYLSTKYSSIFNALFTNAHGESLLAEMGCYGLGVTRILAAAIEVLSTEDC--IR 398
Query 107 FPPQVAPFCVAIIP 120
+P +AP+ V +IP
Sbjct 399 WPRLLAPYQVCVIP 412
> xla:379241 pars2, MGC53152; prolyl-tRNA synthetase 2, mitochondrial
(putative); K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=466
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/76 (38%), Positives = 43/76 (56%), Gaps = 10/76 (13%)
Query 49 LEVGHCFQLPKAYCKAARARFMDSQGVEKVPFMSS---YGLGVSRLIAH--LALSHQDMK 103
+E+GH F L Y +A DSQ K PF++ YG+GVSRL+A LS++D
Sbjct 296 IEIGHTFYLGTKYSHVFKANCYDSQ---KTPFLAEMGCYGIGVSRLLAASLEVLSNED-- 350
Query 104 GLLFPPQVAPFCVAII 119
+ +P +AP+ V +I
Sbjct 351 DIHWPGLIAPYQVCLI 366
> cpv:cgd6_4400 proline-tRNA synthetase; class II aaRS (ybak RNA
binding domain plus tRNA synthetase) ; K01881 prolyl-tRNA
synthetase [EC:6.1.1.15]
Length=719
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 29/101 (28%), Positives = 45/101 (44%), Gaps = 7/101 (6%)
Query 22 TPRLDRTNESHNGAHGATAPYVCGGTCLEVGHCFQ------LPKAYCKAARARFMDSQGV 75
TP + T + G G E+G Q L + + K F D +G
Sbjct 413 TPVVKGTKSENEKFPGGDITKSIEGFIPEIGRAVQAATSHLLGQNFSKMFGVEFEDEKGN 472
Query 76 EKVPFMSSYGLGVSRLIAHLALSHQDMKGLLFPPQVAPFCV 116
++ +S+GL +R I + ++H D KGL+ PP+VAP V
Sbjct 473 KEYAHQTSWGL-TTRAIGVMIMTHGDNKGLVLPPKVAPVQV 512
> hsa:25973 PARS2, DKFZp727A071, MGC14416, MGC19467, MT-PRORS;
prolyl-tRNA synthetase 2, mitochondrial (putative) (EC:6.1.1.15);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=475
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 39/74 (52%), Gaps = 4/74 (5%)
Query 49 LEVGHCFQLPKAYCKAARARFMDSQGVEKVPFMSSYGLGVSRLIAHL--ALSHQDMKGLL 106
+EVGH F L Y A+F + G + M YGLGV+R++A LS +D +
Sbjct 305 IEVGHTFYLGTKYSSIFNAQFTNVCGKPTLAEMGCYGLGVTRILAAAIEVLSTEDC--VR 362
Query 107 FPPQVAPFCVAIIP 120
+P +AP+ +IP
Sbjct 363 WPSLLAPYQACLIP 376
> tgo:TGME49_019850 prolyl-tRNA synthetase, putative (EC:6.1.1.15);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=711
Score = 40.0 bits (92), Expect = 0.003, Method: Composition-based stats.
Identities = 20/60 (33%), Positives = 34/60 (56%), Gaps = 1/60 (1%)
Query 61 YCKAARARFMDSQGVEKVPFMSSYGLGVSRLIAHLALSHQDMKGLLFPPQVAPFCVAIIP 120
+ K F D +G +++ +S+G +R + + ++H D KGL+ PP+VA V IIP
Sbjct 447 FAKMFEIEFEDEEGHKRLVHQTSWGC-TTRSLGVMIMTHGDDKGLVIPPRVASVQVVIIP 505
> pfa:PFL0670c bifunctional aminoacyl-tRNA synthetase, putative
(EC:6.1.1.15); K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=746
Score = 40.0 bits (92), Expect = 0.004, Method: Composition-based stats.
Identities = 21/75 (28%), Positives = 37/75 (49%), Gaps = 1/75 (1%)
Query 46 GTCLEVGHCFQLPKAYCKAARARFMDSQGVEKVPFMSSYGLGVSRLIAHLALSHQDMKGL 105
G ++ L + K + F D V++ +S+G +R I + ++H D KGL
Sbjct 471 GRAIQAATSHYLGTNFAKMFKIEFEDENEVKQYVHQTSWGC-TTRSIGIMIMTHGDDKGL 529
Query 106 LFPPQVAPFCVAIIP 120
+ PP V+ + V I+P
Sbjct 530 VLPPNVSKYKVVIVP 544
> ath:AT3G62120 tRNA synthetase class II (G, H, P and S) family
protein; K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=530
Score = 38.9 bits (89), Expect = 0.009, Method: Composition-based stats.
Identities = 21/70 (30%), Positives = 37/70 (52%), Gaps = 3/70 (4%)
Query 53 HCFQLPKAYCKAARARFMDSQGVEKVPFMSSYGLGVSRLIAHLALSHQDMKGLLFPPQVA 112
HC L + + K F + + ++ + +S+ +R I + ++H D KGL+ PP+VA
Sbjct 275 HC--LGQNFAKMFEINFENEKAETEMVWQNSWAYS-TRTIGVMIMTHGDDKGLVLPPKVA 331
Query 113 PFCVAIIPQP 122
V +IP P
Sbjct 332 SVQVVVIPVP 341
> hsa:2058 EPRS, DKFZp313B047, EARS, GLUPRORS, PARS, QARS, QPRS;
glutamyl-prolyl-tRNA synthetase (EC:6.1.1.15 6.1.1.17); K14163
bifunctional glutamyl/prolyl-tRNA synthetase [EC:6.1.1.17
6.1.1.15]
Length=1512
Score = 38.5 bits (88), Expect = 0.012, Method: Composition-based stats.
Identities = 24/77 (31%), Positives = 39/77 (50%), Gaps = 3/77 (3%)
Query 46 GTCLEVGHCFQLPKAYCKAARARFMDSQ--GVEKVPFMSSYGLGVSRLIAHLALSHQDMK 103
G ++ G L + + K F D + G ++ + +S+GL +R I + + H D
Sbjct 1233 GRAIQGGTSHHLGQNFSKMFEIVFEDPKIPGEKQFAYQNSWGL-TTRTIGVMTMVHGDNM 1291
Query 104 GLLFPPQVAPFCVAIIP 120
GL+ PP+VA V IIP
Sbjct 1292 GLVLPPRVACVQVVIIP 1308
> cel:T20H4.3 prs-1; Prolyl tRNA Synthetase family member (prs-1);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=581
Score = 37.4 bits (85), Expect = 0.025, Method: Composition-based stats.
Identities = 22/79 (27%), Positives = 38/79 (48%), Gaps = 3/79 (3%)
Query 44 CGGTCLEVGHCFQLPKAYCKAARARFMD--SQGVEKVPFMSSYGLGVSRLIAHLALSHQD 101
C G ++ L + + K + D +G + +S+GL +R I + + H D
Sbjct 306 CNGRGIQGATSHHLGQNFSKMFDISYEDPAKEGERAFAWQNSWGLS-TRTIGAMVMIHGD 364
Query 102 MKGLLFPPQVAPFCVAIIP 120
KGL+ PP+VA V ++P
Sbjct 365 DKGLVLPPRVAAVQVIVVP 383
> sce:YHR020W Protein of unknown function that may interact with
ribosomes, based on co-purification experiments; has similarity
to proline-tRNA ligase; YHR020W is an essential gene
(EC:6.1.1.15); K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=688
Score = 37.4 bits (85), Expect = 0.026, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 27/41 (65%), Gaps = 1/41 (2%)
Query 80 FMSSYGLGVSRLIAHLALSHQDMKGLLFPPQVAPFCVAIIP 120
+ +S+GL +R+I + + H D KGL+ PP+V+ F +IP
Sbjct 441 YQNSWGLS-TRVIGVMVMIHSDNKGLVIPPRVSQFQSVVIP 480
> cel:T27F6.5 prs-2; Prolyl tRNA Synthetase family member (prs-2);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=454
Score = 37.0 bits (84), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 40/80 (50%), Gaps = 6/80 (7%)
Query 49 LEVGHCFQLPKAYCKAARARFMDSQGVEKVPFMSSYGLGVSRLI-AHLALSHQDMKGLLF 107
+E+ H F L Y +A A+F QG K M +G+GV+RL+ A + L K L
Sbjct 262 VEIAHTFHLGTKYSEALGAKF---QG--KPLDMCCFGIGVTRLLPAAIDLLSVSDKALRL 316
Query 108 PPQVAPFCVAIIPQPAARWN 127
P +APF II + + N
Sbjct 317 PRAIAPFDAVIIVKKSLMSN 336
> xla:100158287 hypothetical protein LOC100158287; K01881 prolyl-tRNA
synthetase [EC:6.1.1.15]
Length=655
Score = 36.6 bits (83), Expect = 0.036, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 30/49 (61%), Gaps = 1/49 (2%)
Query 72 SQGVEKVPFMSSYGLGVSRLIAHLALSHQDMKGLLFPPQVAPFCVAIIP 120
+ G+++ + +S+GL +R I + + H D G++ PP+VA V IIP
Sbjct 404 TPGLKQYAYQNSWGLS-TRTIGAMVMVHADNIGMVLPPRVACVQVVIIP 451
> xla:100158438 eprs; glutamyl-prolyl-tRNA synthetase; K14163
bifunctional glutamyl/prolyl-tRNA synthetase [EC:6.1.1.17 6.1.1.15]
Length=1499
Score = 36.6 bits (83), Expect = 0.040, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 30/49 (61%), Gaps = 1/49 (2%)
Query 72 SQGVEKVPFMSSYGLGVSRLIAHLALSHQDMKGLLFPPQVAPFCVAIIP 120
+ G+++ + +S+GL +R I + + H D G++ PP+VA + IIP
Sbjct 1248 TPGLKQYAYQNSWGLS-TRTIGAMVMVHADNTGMVLPPRVACVQIIIIP 1295
> dre:562037 eprs, wu:fb38d08, wu:ft34d10; glutamyl-prolyl-tRNA
synthetase; K14163 bifunctional glutamyl/prolyl-tRNA synthetase
[EC:6.1.1.17 6.1.1.15]
Length=1511
Score = 36.6 bits (83), Expect = 0.043, Method: Composition-based stats.
Identities = 18/47 (38%), Positives = 29/47 (61%), Gaps = 1/47 (2%)
Query 74 GVEKVPFMSSYGLGVSRLIAHLALSHQDMKGLLFPPQVAPFCVAIIP 120
G +++ + +S+G+ +R I L + H D GL+ PP+VA V IIP
Sbjct 1262 GEKQLAYQNSWGI-TTRTIGVLTMVHGDNMGLVLPPRVACLQVIIIP 1307
> mmu:107508 Eprs, 2410081F06Rik, 3010002K18Rik, C79379, Qprs;
glutamyl-prolyl-tRNA synthetase (EC:6.1.1.15 6.1.1.17); K14163
bifunctional glutamyl/prolyl-tRNA synthetase [EC:6.1.1.17
6.1.1.15]
Length=1512
Score = 36.2 bits (82), Expect = 0.046, Method: Composition-based stats.
Identities = 22/77 (28%), Positives = 37/77 (48%), Gaps = 3/77 (3%)
Query 46 GTCLEVGHCFQLPKAYCKAARARFMDSQ--GVEKVPFMSSYGLGVSRLIAHLALSHQDMK 103
G ++ L + + K F D + G ++ + S+GL +R I + + H D
Sbjct 1233 GRAIQGATSHHLGQNFSKMCEIVFEDPKTPGEKQFAYQCSWGL-TTRTIGVMVMVHGDNM 1291
Query 104 GLLFPPQVAPFCVAIIP 120
GL+ PP+VA V +IP
Sbjct 1292 GLVLPPRVASVQVVVIP 1308
> bbo:BBOV_IV001390 21.m02889; prolyl-tRNA synthetase (EC:6.1.1.15);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=705
Score = 36.2 bits (82), Expect = 0.055, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 30/52 (57%), Gaps = 1/52 (1%)
Query 69 FMDSQGVEKVPFMSSYGLGVSRLIAHLALSHQDMKGLLFPPQVAPFCVAIIP 120
F D GV+++ +S+G +R I + H D +GL+ PP+V+ + I+P
Sbjct 456 FEDENGVKQLAHQTSWGF-TTRSIGVSIMIHGDDQGLVIPPRVSKVQIVIVP 506
> tpv:TP01_0166 prolyl-tRNA synthetase; K01881 prolyl-tRNA synthetase
[EC:6.1.1.15]
Length=683
Score = 35.8 bits (81), Expect = 0.075, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 31/53 (58%), Gaps = 1/53 (1%)
Query 68 RFMDSQGVEKVPFMSSYGLGVSRLIAHLALSHQDMKGLLFPPQVAPFCVAIIP 120
++ D +G ++ +S+G +R I + + H D +GL+ PP+VA V I+P
Sbjct 440 KYEDEEGEKRKVHQTSWGF-TTRSIGIMIMVHGDDRGLVVPPRVAKTQVVIVP 491
> ath:AT5G10880 tRNA synthetase-related / tRNA ligase-related
Length=309
Score = 35.0 bits (79), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 14/26 (53%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 97 LSHQDMKGLLFPPQVAPFCVAIIPQP 122
++H D KGL+FPP+VAP V +I P
Sbjct 89 MTHGDDKGLVFPPKVAPVQVVVIHVP 114
> pfa:PF10_0183 eukaryotic translation initiation factor subunit
eIF2A, putative
Length=2405
Score = 32.0 bits (71), Expect = 0.93, Method: Composition-based stats.
Identities = 11/23 (47%), Positives = 14/23 (60%), Gaps = 0/23 (0%)
Query 106 LFPPQVAPFCVAIIPQPAARWND 128
LFPP + PF V PQP +W +
Sbjct 2380 LFPPPLKPFTVVPKPQPVLQWEE 2402
Lambda K H
0.328 0.138 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4027755848
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40