bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0921_orf1
Length=100
Score E
Sequences producing significant alignments: (Bits) Value
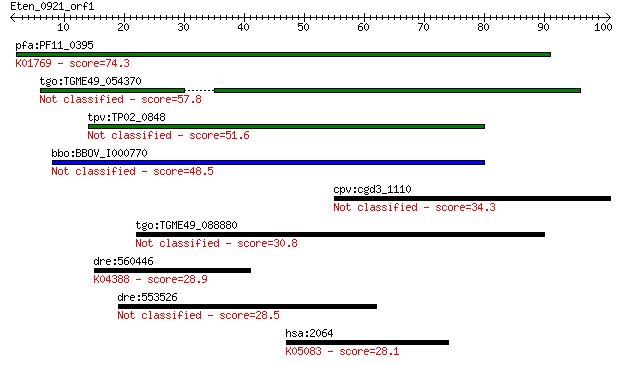
pfa:PF11_0395 guanylyl cyclase; K01769 guanylate cyclase, othe... 74.3 9e-14
tgo:TGME49_054370 adenylate and guanylate cyclase catalytic do... 57.8 8e-09
tpv:TP02_0848 guanylyl cyclase 51.6 7e-07
bbo:BBOV_I000770 16.m00775; adenylate and guanylate cyclase ca... 48.5 6e-06
cpv:cgd3_1110 P-type ATpase fused to two adenyl cyclase domain... 34.3 0.10
tgo:TGME49_088880 hypothetical protein 30.8 1.2
dre:560446 similar to TGF-beta type II receptor; K04388 TGF-be... 28.9 4.4
dre:553526 MGC152774, ZCWCC3; zgc:152774 28.5 5.1
hsa:2064 ERBB2, CD340, HER-2, HER-2/neu, HER2, MLN_19, NEU, NG... 28.1 6.4
> pfa:PF11_0395 guanylyl cyclase; K01769 guanylate cyclase, other
[EC:4.6.1.2]
Length=4226
Score = 74.3 bits (181), Expect = 9e-14, Method: Composition-based stats.
Identities = 39/95 (41%), Positives = 58/95 (61%), Gaps = 6/95 (6%)
Query 2 SWMGIRWLIFWLNILFIFAA----LAFVLTNARDPGKKAPSEWHTADTVELSFFLTLIHH 57
S + I+W+IF+LN+LFI AA +A++ + + + W T DT+E F+L ++HH
Sbjct 3804 SLLNIKWMIFFLNLLFISAACVFSIAYLWAISETDQTTSYTIWMTNDTIEFFFYLVILHH 3863
Query 58 NTGLLFQHIIFVDALLITAS--FSSAMLVSQPTTE 90
NTG+LFQ I VD L IT S F + +V TT+
Sbjct 3864 NTGMLFQTCILVDLLFITMSLTFIATSVVKTITTD 3898
> tgo:TGME49_054370 adenylate and guanylate cyclase catalytic
domain-containing protein (EC:4.6.1.1 3.6.3.1 1.15.1.1 4.6.1.2
3.1.3.48 2.8.3.8)
Length=4368
Score = 57.8 bits (138), Expect = 8e-09, Method: Composition-based stats.
Identities = 25/61 (40%), Positives = 40/61 (65%), Gaps = 0/61 (0%)
Query 35 KAPSEWHTADTVELSFFLTLIHHNTGLLFQHIIFVDALLITASFSSAMLVSQPTTERLRG 94
+A + W +DT+EL F++ ++HHNTGLLFQ+ I VD LL+T S + + ++ T +
Sbjct 3897 RAYTYWLLSDTIELFFYIVILHHNTGLLFQNCILVDVLLMTMSLTFIITTARETASTVST 3956
Query 95 I 95
I
Sbjct 3957 I 3957
Score = 34.3 bits (77), Expect = 0.11, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 19/24 (79%), Gaps = 0/24 (0%)
Query 6 IRWLIFWLNILFIFAALAFVLTNA 29
+RW++F LN+LFI A+ F L+N+
Sbjct 3773 LRWMVFLLNLLFISASCVFALSNS 3796
> tpv:TP02_0848 guanylyl cyclase
Length=2664
Score = 51.6 bits (122), Expect = 7e-07, Method: Composition-based stats.
Identities = 23/66 (34%), Positives = 39/66 (59%), Gaps = 0/66 (0%)
Query 14 NILFIFAALAFVLTNARDPGKKAPSEWHTADTVELSFFLTLIHHNTGLLFQHIIFVDALL 73
N LF L ++LT + + W +D+VE F+L L+HH+TG+LFQ+ + +D+L
Sbjct 2311 NYLFQLTNLLYILTLCHNGCSVDYNLWLNSDSVEFYFYLILLHHSTGMLFQNCLLIDSLF 2370
Query 74 ITASFS 79
+ S +
Sbjct 2371 LVLSMT 2376
> bbo:BBOV_I000770 16.m00775; adenylate and guanylate cyclase
catalytic domain containing protein
Length=2446
Score = 48.5 bits (114), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 26/77 (33%), Positives = 40/77 (51%), Gaps = 5/77 (6%)
Query 8 WLIFWLNILFIFAALAFVLTNA-----RDPGKKAPSEWHTADTVELSFFLTLIHHNTGLL 62
WL F LN+ F+ AA F L+ + P + W +D + ++ +IHHN G+L
Sbjct 2095 WLTFALNLFFVSAACVFSLSGSWAVEHNHPNFHLANIWLPSDNFKFYTYIVVIHHNNGML 2154
Query 63 FQHIIFVDALLITASFS 79
FQ + VD+L + S S
Sbjct 2155 FQTCLLVDSLFMVISMS 2171
> cpv:cgd3_1110 P-type ATpase fused to two adenyl cyclase domains
and 21 predicted transmembrane regions
Length=3848
Score = 34.3 bits (77), Expect = 0.10, Method: Composition-based stats.
Identities = 19/49 (38%), Positives = 28/49 (57%), Gaps = 9/49 (18%)
Query 55 IHHNTGLLFQHIIFVDAL---LITASFSSAMLVSQPTTERLRGILIVCL 100
+HHN GLLF++I+ D L LI FS + V+ L GI++ C+
Sbjct 3424 VHHNAGLLFKYIVLFDLLIMFLILTIFSVGINVN------LEGIVVYCV 3466
> tgo:TGME49_088880 hypothetical protein
Length=599
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 21/85 (24%), Positives = 35/85 (41%), Gaps = 17/85 (20%)
Query 22 LAFVLTNARDPGKKAPSEWH-----------------TADTVELSFFLTLIHHNTGLLFQ 64
L+ +L AR+PG K S H AD +E++ L+ +H +G
Sbjct 420 LSLLLATAREPGGKCHSRLHFPRNSRGAAALSGLCLAVADEIEVNLALSWVHLLSGFARA 479
Query 65 HIIFVDALLITASFSSAMLVSQPTT 89
HI D + A S+ L ++ +
Sbjct 480 HIAHADLFEVAALPLSSFLETKAAS 504
> dre:560446 similar to TGF-beta type II receptor; K04388 TGF-beta
receptor type-2 [EC:2.7.11.30]
Length=576
Score = 28.9 bits (63), Expect = 4.4, Method: Composition-based stats.
Identities = 12/26 (46%), Positives = 16/26 (61%), Gaps = 1/26 (3%)
Query 15 ILFIFAALAFVLTNARDPGKKAPSEW 40
++ + A +AF L R PGKK P EW
Sbjct 142 LVAVIATMAFYLYRTRQPGKK-PKEW 166
> dre:553526 MGC152774, ZCWCC3; zgc:152774
Length=306
Score = 28.5 bits (62), Expect = 5.1, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 23/43 (53%), Gaps = 0/43 (0%)
Query 19 FAALAFVLTNARDPGKKAPSEWHTADTVELSFFLTLIHHNTGL 61
F+A+A ++ NA DPG A + W TV L+ + +G+
Sbjct 30 FSAVAELIDNASDPGVTAKNIWIDVVTVRDQLCLSFTDNGSGM 72
> hsa:2064 ERBB2, CD340, HER-2, HER-2/neu, HER2, MLN_19, NEU,
NGL, TKR1; v-erb-b2 erythroblastic leukemia viral oncogene homolog
2, neuro/glioblastoma derived oncogene homolog (avian)
(EC:2.7.10.1); K05083 receptor tyrosine-protein kinase erbB-2
[EC:2.7.10.1]
Length=1225
Score = 28.1 bits (61), Expect = 6.4, Method: Composition-based stats.
Identities = 14/27 (51%), Positives = 15/27 (55%), Gaps = 0/27 (0%)
Query 47 ELSFFLTLIHHNTGLLFQHIIFVDALL 73
EL L LIHHNT L F H + D L
Sbjct 430 ELGSGLALIHHNTHLCFVHTVPWDQLF 456
Lambda K H
0.332 0.141 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2041372988
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40